Colonoscopy
Introduction
In this section we will:
* identify which colonoscopy procedures were excluded and why
* identify which different colonoscopy subgroups were created and why
If you have questions or concerns about this data please contact Alexander Nielson (alexnielson@utah.gov)
Load Libraries
Load Libraries
library(data.table)
library(tidyverse)## Warning: replacing previous import 'vctrs::data_frame' by 'tibble::data_frame'
## when loading 'dplyr'library(stringi)
library(ggridges)
library(broom)
library(disk.frame)
library(RecordLinkage)
library(googlesheets4)
library(bigrquery)
library(DBI)
devtools::install_github("utah-osa/hcctools2", upgrade="never" )
library(hcctools2)Establish color palettes
cust_color_pal1 <- c(
"Anesthesia" = "#f3476f",
"Facility" = "#e86a33",
"Medicine Proc" = "#e0a426",
"Pathology" = "#77bf45",
"Radiology" = "#617ff7",
"Surgery" = "#a974ee"
)
cust_color_pal2 <- c(
"TRUE" = "#617ff7",
"FALSE" = "#e0a426"
)
cust_color_pal3 <- c(
"above avg" = "#f3476f",
"avg" = "#77bf45",
"below avg" = "#a974ee"
)
fac_ref_regex <- "(UTAH)|(IHC)|(HOSP)|(HOSPITAL)|(CLINIC)|(ANESTH)|(SCOPY)|(SURG)|(LLC)|(ASSOC)|(MEDIC)|(CENTER)|(ASSOCIATES)|(U OF U)|(HEALTH)|(OLOGY)|(OSCOPY)|(FAMILY)|(VAMC)|(SLC)|(SALT LAKE)|(CITY)|(PROVO)|(OGDEN)|(ENDO )|( VALLEY)|( REGIONAL)|( CTR)|(GRANITE)|( INSTITUTE)|(INSTACARE)|(WASATCH)|(COUNTY)|(PEDIATRIC)|(CORP)|(CENTRAL)|(URGENT)|(CARE)|(UNIV)|(ODYSSEY)|(MOUNTAINSTAR)|( ORTHOPEDIC)|(INSTITUT)|(PARTNERSHIP)|(PHYSICIAN)|(CASTLEVIEW)|(CONSULTING)|(MAGEMENT)|(PRACTICE)|(EMERGENCY)|(SPECIALISTS)|(DIVISION)|(GUT WHISPERER)|(INTERMOUNTAIN)|(OBGYN)"Connect to GCP database
bigrquery::bq_auth(path = 'D:/gcp_keys/healthcare-compare-prod-95b3b7349c32.json')
# set my project ID and dataset name
project_id <- 'healthcare-compare-prod'
dataset_name <- 'healthcare_compare'
con <- dbConnect(
bigrquery::bigquery(),
project = project_id,
dataset = dataset_name,
billing = project_id
)Get NPI table
query <- paste0("SELECT npi, clean_name, osa_group, osa_class, osa_specialization
FROM `healthcare-compare-prod.healthcare_compare.npi_full`")
#bq_project_query(billing, query) # uncomment to determine billing price for above query.
npi_full <- dbGetQuery(con, query) %>%
data.table() get a subset of the NPI providers based upon taxonomy groups
gs4_auth(email="alexnielson@utah.gov")surgery <- read_sheet("https://docs.google.com/spreadsheets/d/1GY8lKwUJuPHtpUl4EOw9eriLUDG9KkNWrMbaSnA5hOU/edit#gid=0",
sheet="major_surgery") %>% as.data.table## Reading from "Doctor Types to Keep"## Range "'major_surgery'"surgery <- surgery[is.na(Remove) ] %>% .[["NUCC Classification"]] npi_prov_pair <- npi_full[osa_class %in% surgery] %>%
.[,.(npi=npi,
clean_name = clean_name
)
] Load Data
bun_proc <- disk.frame("full_apcd.df")cscopy <- bun_proc[surg_bun_t_colonoscopy == T & cnt >5 & tp_med < 10000]cscopy <- cscopy[,`:=`(
surg_sp_name_clean = surg_sp_npi %>% map_chr(get_npi_standard_name),
surg_bp_name_clean = surg_bp_npi %>% map_chr(get_npi_standard_name),
medi_sp_name_clean = medi_sp_npi %>% map_chr(get_npi_standard_name),
medi_bp_name_clean = medi_bp_npi %>% map_chr(get_npi_standard_name),
radi_sp_name_clean = radi_sp_npi %>% map_chr(get_npi_standard_name),
radi_bp_name_clean = radi_bp_npi %>% map_chr(get_npi_standard_name),
path_sp_name_clean = path_sp_npi %>% map_chr(get_npi_standard_name),
path_bp_name_clean = path_bp_npi %>% map_chr(get_npi_standard_name),
anes_sp_name_clean = anes_sp_npi %>% map_chr(get_npi_standard_name),
anes_bp_name_clean = anes_bp_npi %>% map_chr(get_npi_standard_name),
faci_sp_name_clean = faci_sp_npi %>% map_chr(get_npi_standard_name),
faci_bp_name_clean = faci_bp_npi %>% map_chr(get_npi_standard_name)
)]cscopy %>% saveRDS("cscopy.RDS")cscopy <- readRDS("cscopy.RDS")anything under 600 is known to be too low. while some clinics may perform colonoscopies at a lower rate, a 600 dolar charge is not typical.
cscopy <- cscopy[tp_med > 600 & tp_med < 10000]cscopy[,tp_med] %>% summary()## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 605.3 1709.2 2158.0 2624.4 3170.9 9996.3A. Standard
We want to find the typical/standard colonoscopy. To do this we need to split and filter out the procedures that we would not typically think as a standard colonoscopy. These groups may be valid and deserve their own category, or they may be rare procedure types which we do not believe the average customer is shopping for.
cscopy %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
cscopy %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 510 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_egd 0.374
## 2 surg_bun_t_tip 0.371
## 3 surg_bun_t_biopsy 0.268
## 4 path_bun_t_patho 0.256
## 5 path_bun_t_pathologist 0.256
## 6 path_bun_t_tissue 0.256
## 7 medi_bun_t_sodium 0.238
## 8 surg_bun_t_sub 0.212
## 9 surg_bun_t_njx 0.208
## 10 path_bun_t_immunohisto 0.193
## # ... with 500 more rowslets compare the procedures which have an EGD and a colonoscopy
cscopy %>% get_tag_density_information(tag="surg_bun_t_egd") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_egd"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 172## $dist_plots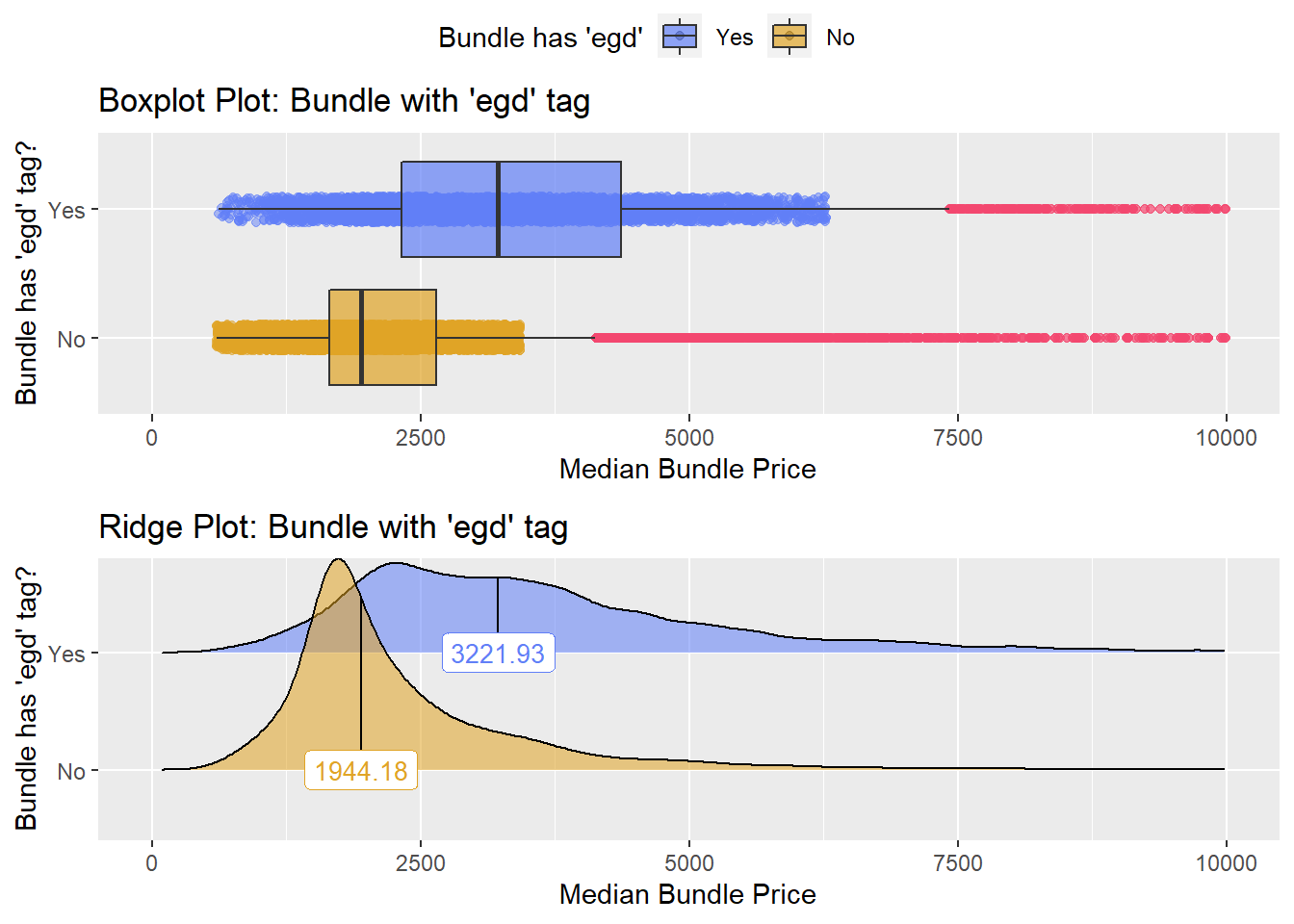
##
## $stat_tables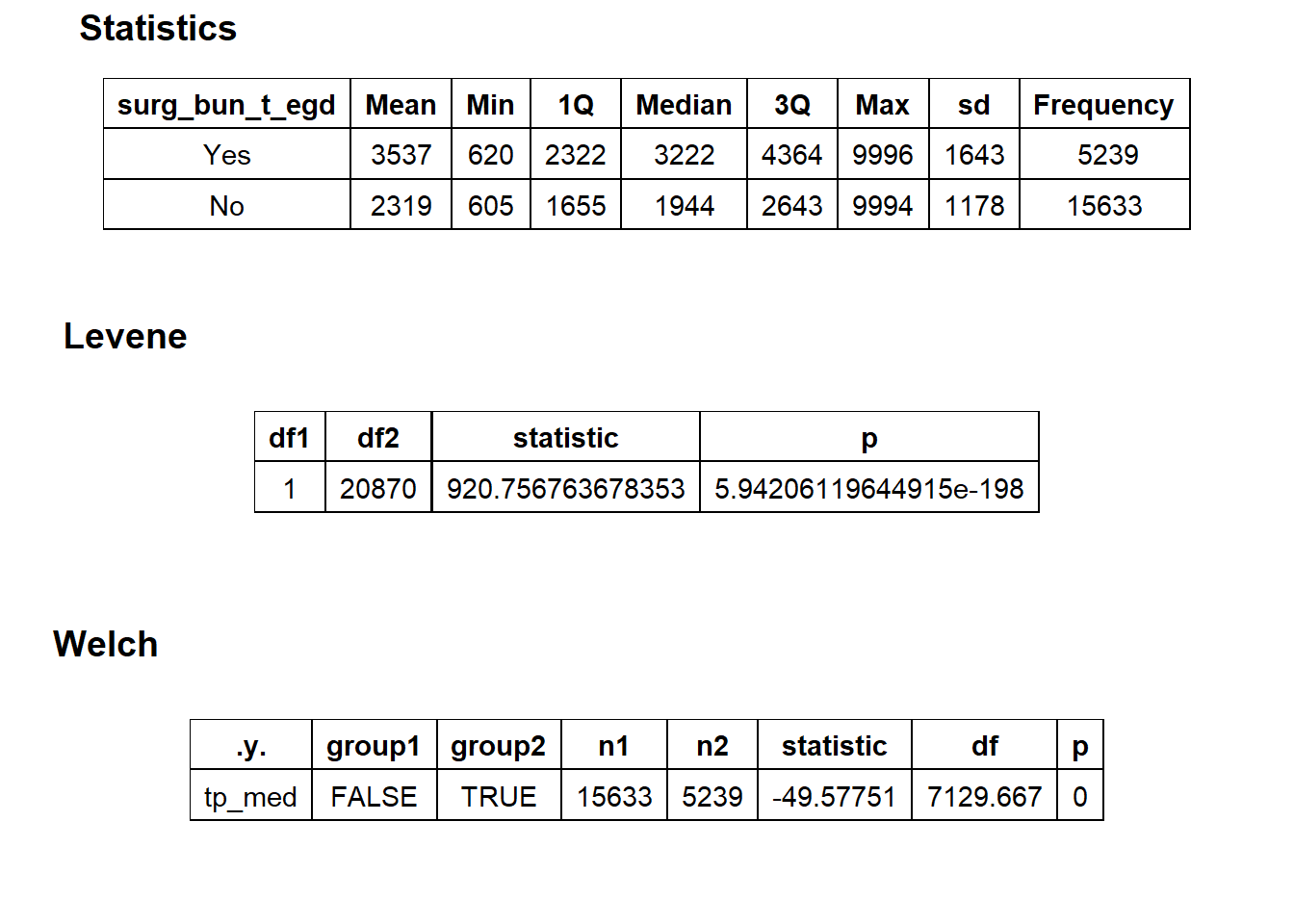
these are clearly different. If a patient has an EGD and a colonoscopy, they are being charged much more than if there was no EGD.
cscopy_with_egd <- cscopy[surg_bun_t_egd ==T]
cscopy <- cscopy[surg_bun_t_egd ==F]Lets now once again examine the distribution and examine any correlated tags
cscopy %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.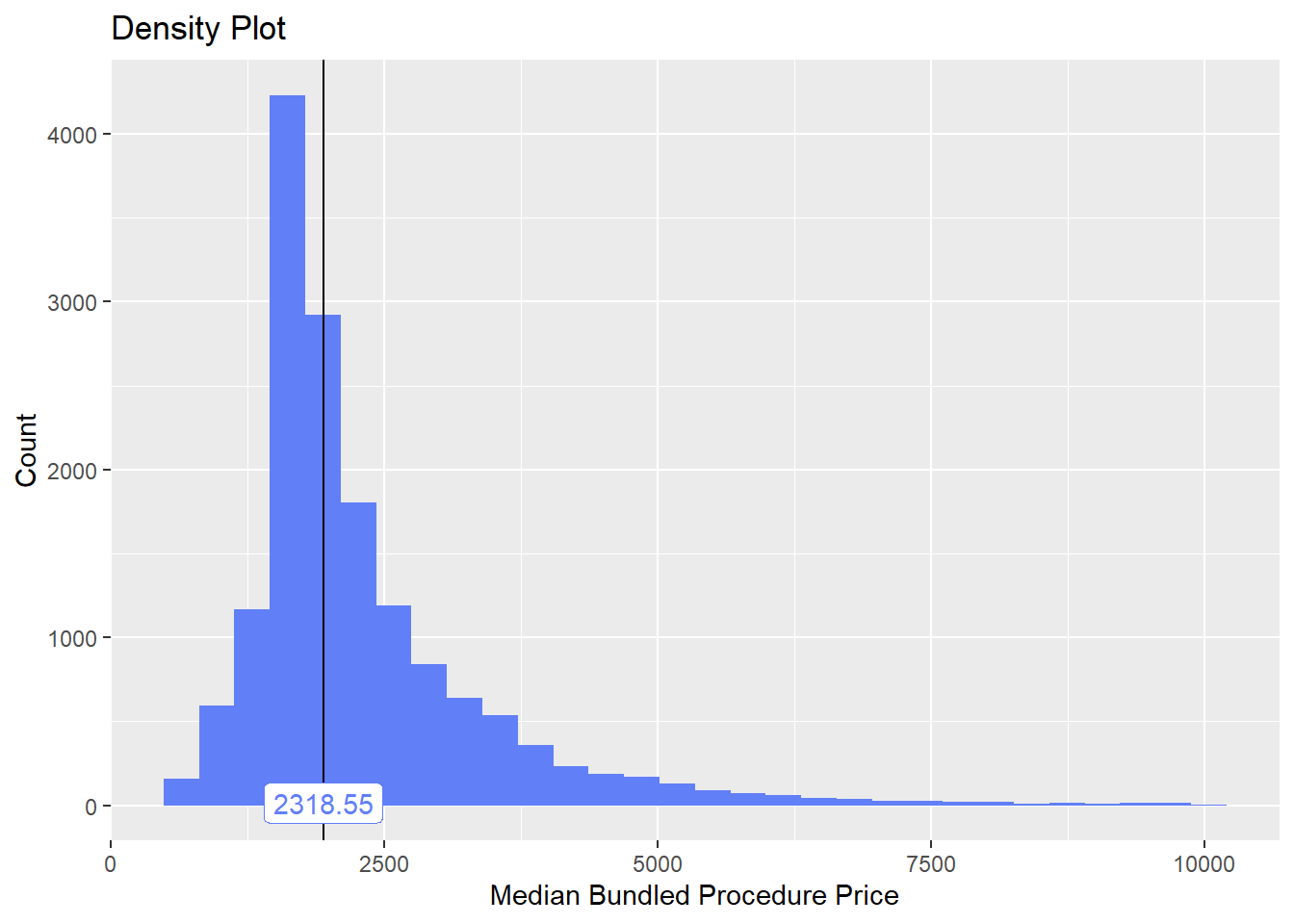
cscopy %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 467 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_sub 0.295
## 2 surg_bun_t_remov 0.291
## 3 surg_bun_t_njx 0.290
## 4 surg_bun_t_removal 0.282
## 5 surg_bun_t_lesion 0.278
## 6 path_bun_t_patho 0.232
## 7 path_bun_t_pathologist 0.232
## 8 path_bun_t_tissue 0.232
## 9 surg_bun_t_diagnostic 0.221
## 10 anes_bun_t_mod_sed 0.202
## # ... with 457 more rowsnow lets examine the colonoscopies with lesion removal
cscopy %>% get_tag_density_information(tag="surg_bun_t_lesion") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_lesion"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 139## $dist_plots
##
## $stat_tables There is strong evidence to remove split these procedures into separate groups. While the Doctor and/or patient may not know if a lesion will be removed prior to surgery, there is a price difference. This will help with post procedure customer analyis.
There is strong evidence to remove split these procedures into separate groups. While the Doctor and/or patient may not know if a lesion will be removed prior to surgery, there is a price difference. This will help with post procedure customer analyis.
cscopy_with_lesion <- cscopy[surg_bun_t_lesion ==T]
cscopy <- cscopy[surg_bun_t_lesion ==F]check distribution and correlated tags
cscopy %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.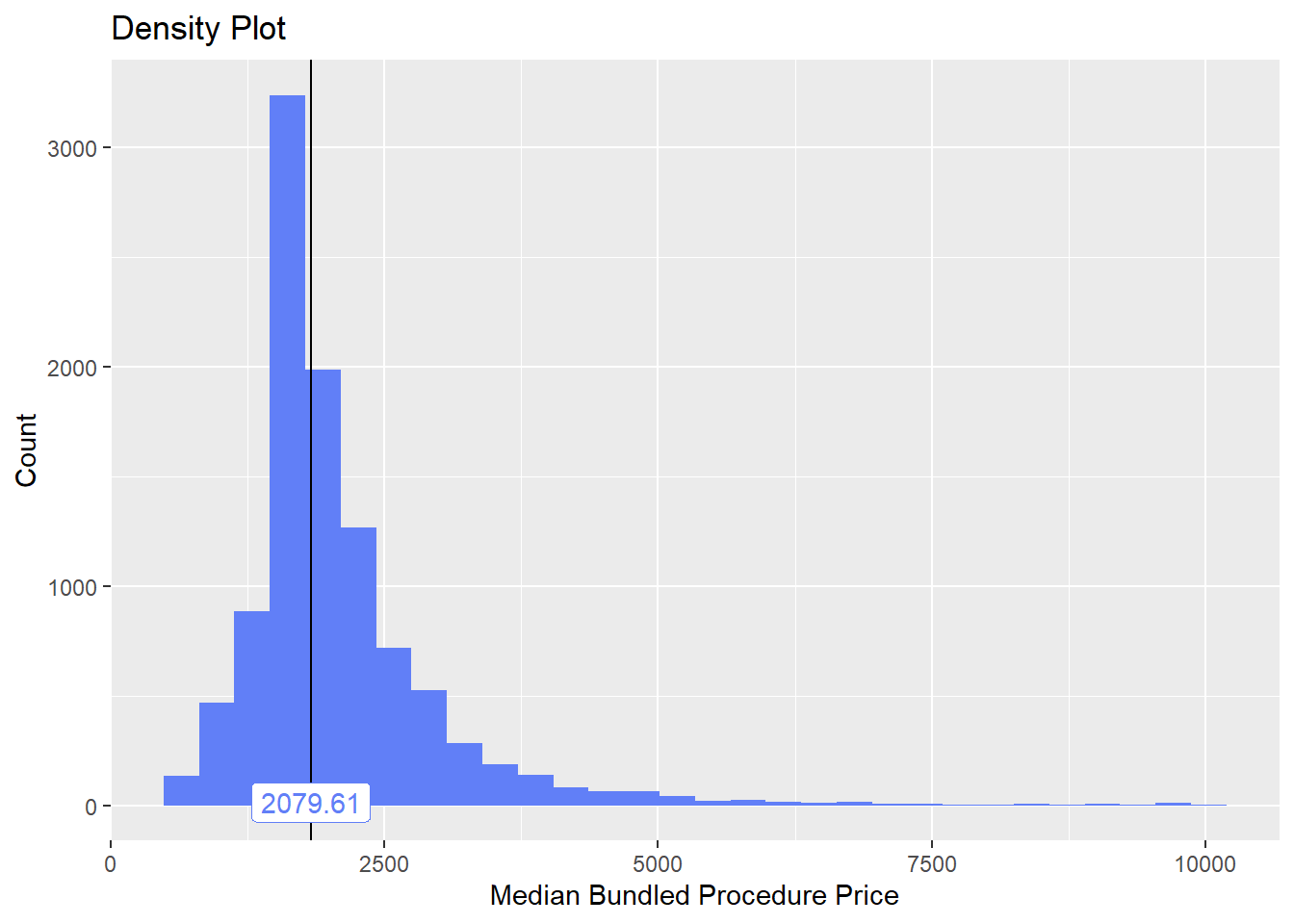
cscopy %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 427 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_anesth 0.257
## 2 anes_bun_t_surg 0.240
## 3 medi_bun_t_sodium 0.212
## 4 anes_bun_t_anes 0.201
## 5 path_bun_t_patho 0.190
## 6 path_bun_t_pathologist 0.190
## 7 path_bun_t_tissue 0.190
## 8 faci_bun_t_dept 0.182
## 9 faci_bun_t_emergency 0.182
## 10 surg_bun_t_remov 0.182
## # ... with 417 more rowscscopy %>% get_tag_density_information("medi_bun_t_inject") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_inject"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 88.1## $dist_plots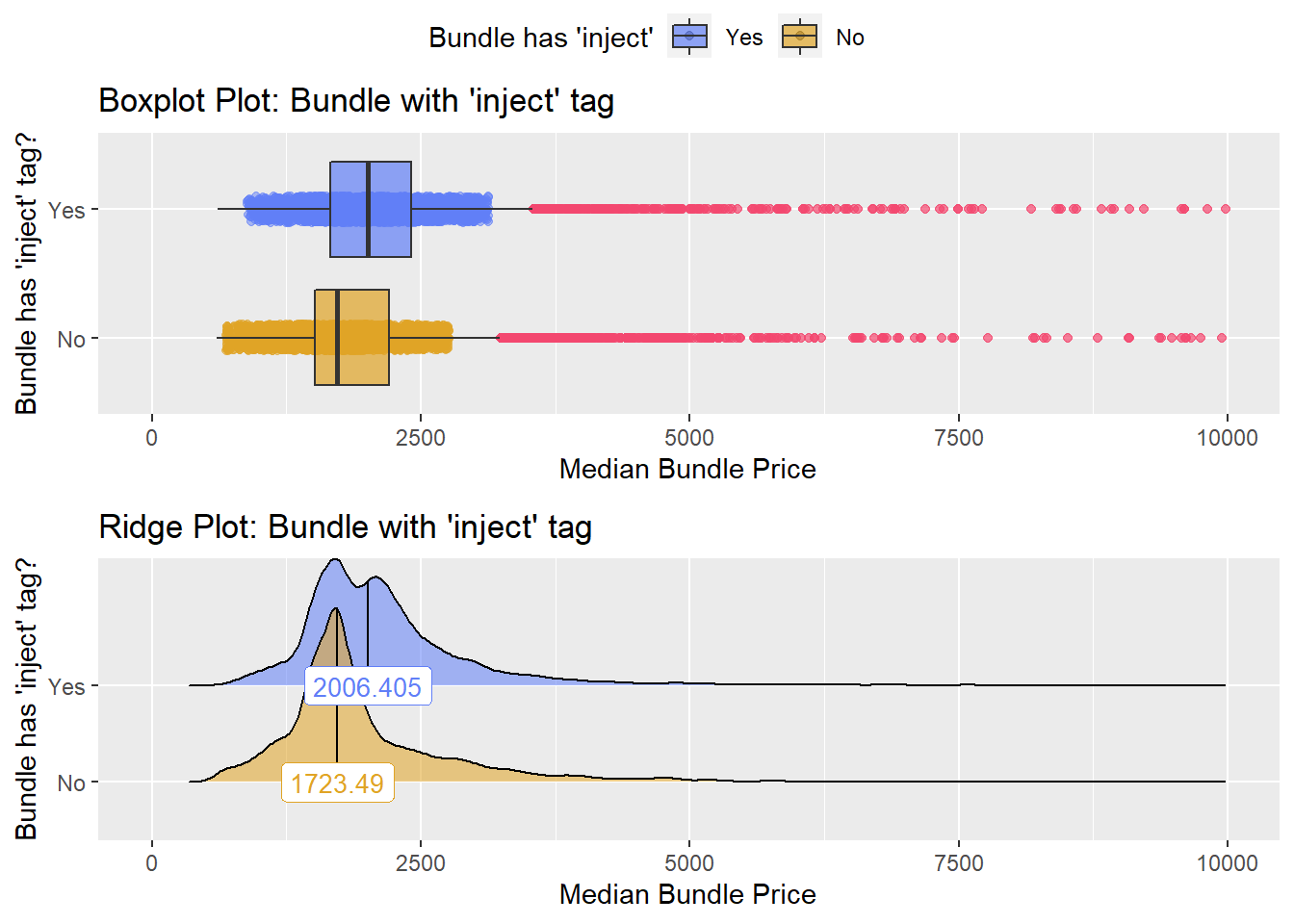
##
## $stat_tables
weak evidence to split based on injection, so for now we will not split. Lets check the tissue pathology next
cscopy %>% get_tag_density_information(tag="path_bun_t_tissue") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_tissue"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 78.3## $dist_plots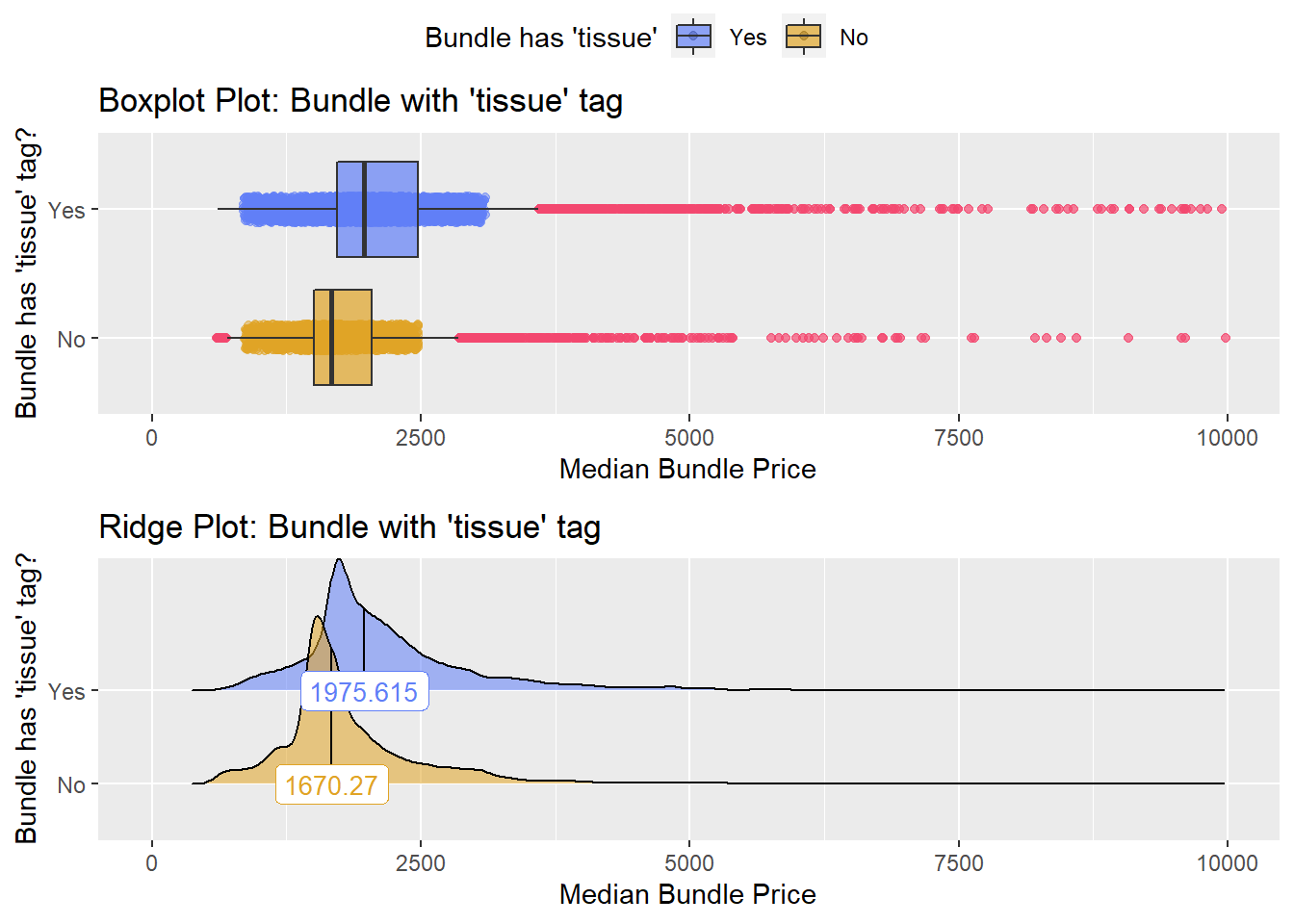
##
## $stat_tables
There is strong evidence to split based upon whether a tissue pathology was conducted. This also passes a gut check since if a tissue had a pathology procedure that it would have cost more to the patient.
cscopy_with_tissue<- cscopy[path_bun_t_tissue ==T]
cscopy <- cscopy[path_bun_t_tissue ==F]cscopy %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.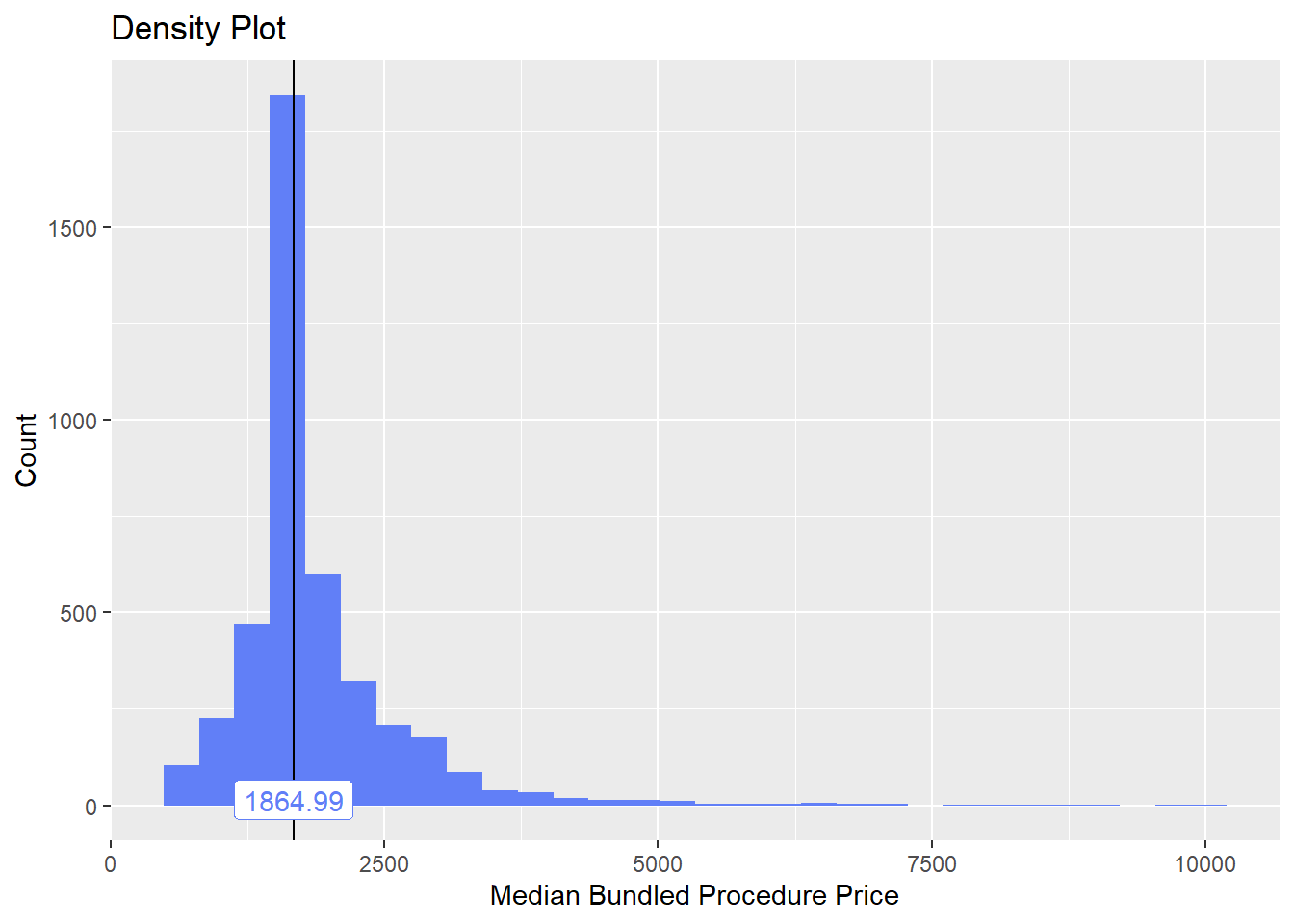
cscopy %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 342 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_dept 0.274
## 2 faci_bun_t_emergency 0.274
## 3 anes_bun_t_anesth 0.251
## 4 anes_bun_t_surg 0.225
## 5 anes_bun_t_anes 0.224
## 6 faci_bun_t_discharge 0.212
## 7 faci_bun_t_initial 0.190
## 8 faci_bun_t_care 0.187
## 9 faci_bun_t_observation 0.186
## 10 surg_bun_t_remov 0.184
## # ... with 332 more rowslets check for colonoscopies at the emergency department
cscopy %>% get_tag_density_information(tag="faci_bun_t_emergency") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ faci_bun_t_emergency"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 581## $dist_plots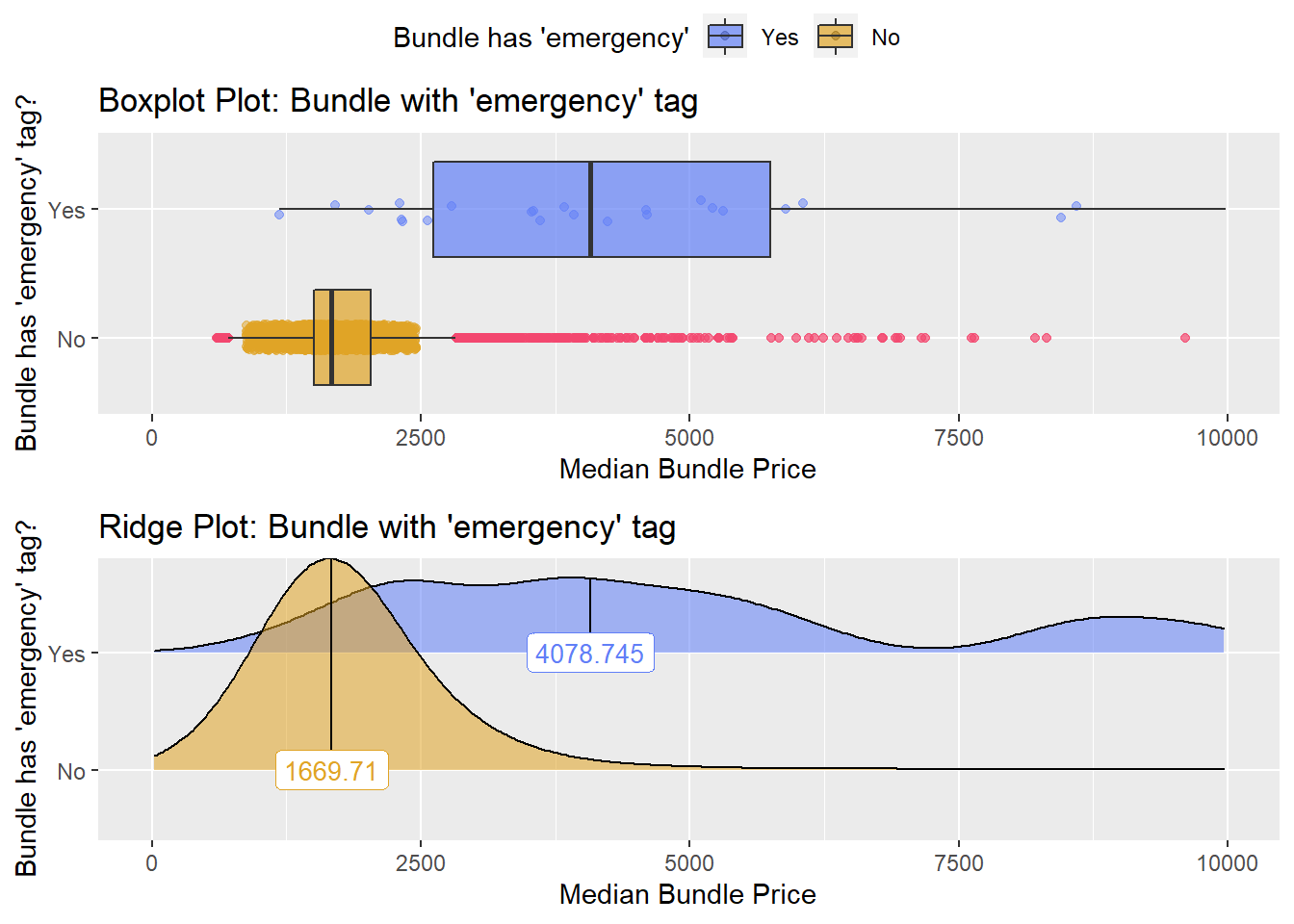
##
## $stat_tables There is strong evidence that a colonoscopy at the emergency department costs much more than one not at an emergency department, however the frequency is not high, so it will not be a serparate searchable category.
There is strong evidence that a colonoscopy at the emergency department costs much more than one not at an emergency department, however the frequency is not high, so it will not be a serparate searchable category.
cscopy_at_emerg <- cscopy[faci_bun_t_emergency==T]
cscopy <- cscopy[faci_bun_t_emergency==F]cscopy %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
cscopy %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 333 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_anesth 0.271
## 2 anes_bun_t_anes 0.250
## 3 anes_bun_t_surg 0.243
## 4 surg_bun_t_remov 0.2
## 5 surg_bun_t_anal 0.194
## 6 surg_bun_t_remove 0.184
## 7 medi_bun_t_sodium 0.180
## 8 surg_bun_t_repair 0.156
## 9 surg_bun_t_complex 0.147
## 10 surg_bun_t_tags 0.140
## # ... with 323 more rowslets remove some obviously non standard additional procedures
cscopy <- cscopy[surg_bun_t_anal==F & surg_bun_t_finger==F & anes_bun_t_hernia==F & surg_bun_t_pharynx==F & surg_bun_t_hernia ==F & surg_bun_t_anal==F & surg_bun_t_lap==F ]cscopy %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.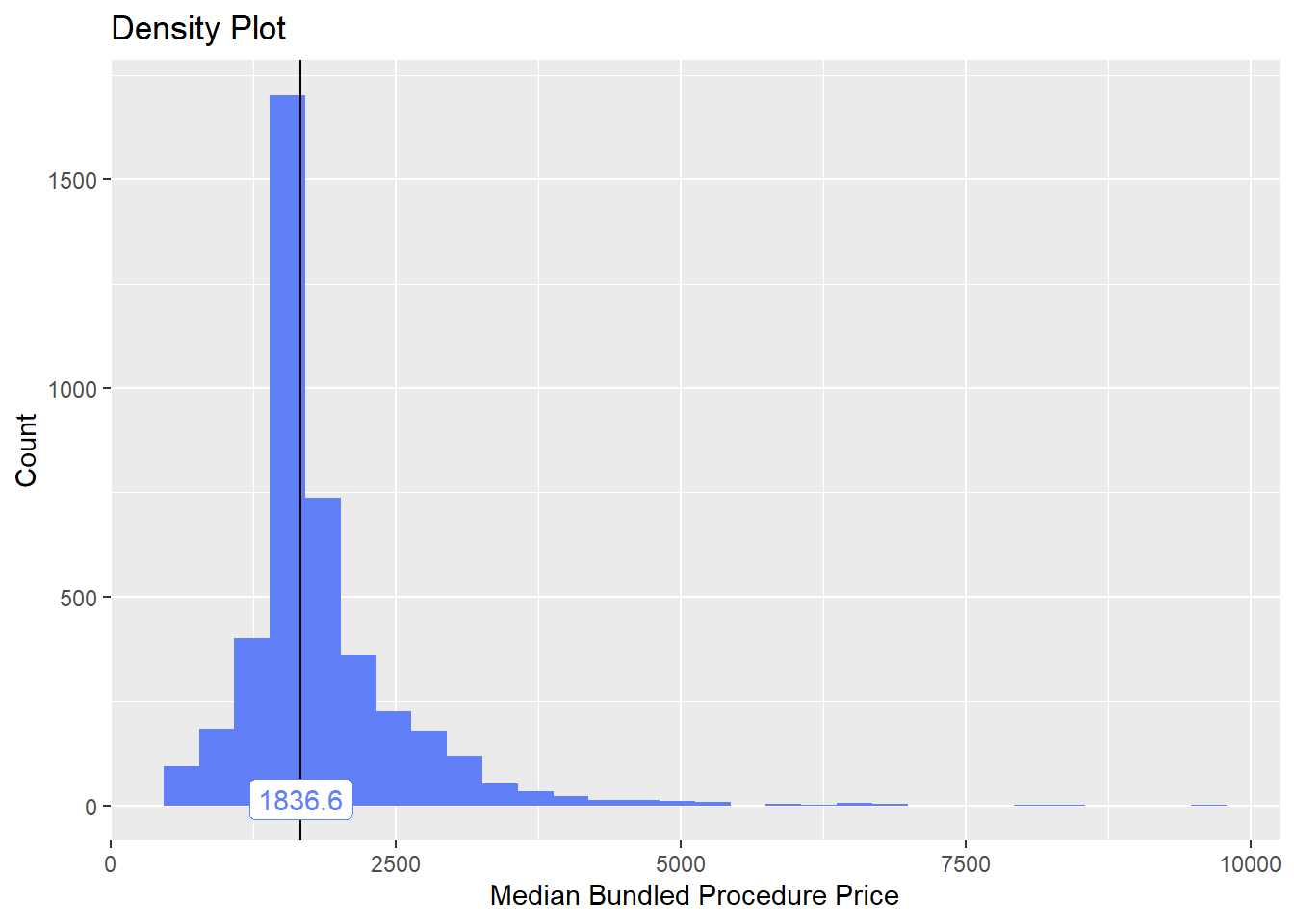 We are now happy with this distribution and values for the standard colonoscopy.
Now we will group the bundles by the facility, and doctor pairs based upon the NPI database.
We are now happy with this distribution and values for the standard colonoscopy.
Now we will group the bundles by the facility, and doctor pairs based upon the NPI database.
cscopy_btbv4 <- cscopy %>% btbv4()For colonoscopies, we will only show those which performed at least 25 surgeries.
cscopy_btbv4 <- cscopy_btbv4 %>%
arrange(most_important_fac) %>%
filter(tp_cnt_cnt>9) %>%
select(doctor_str1,
doctor_str2,
doctor_npi_str1,
doctor_npi_str2,
most_important_fac,
most_important_fac_npi,
tp_med_med,
tp_med_surg,
tp_med_medi,
tp_med_path,
tp_med_radi,
tp_med_anes,
tp_med_faci,
tp_cnt_cnt) %>%
mutate(procedure_type = 1,
procedure_modifier = "Standard")cscopy_btbv4 %>% as.data.frame()%>% count(doctor_str1,doctor_str2, most_important_fac) %>% filter(n>1)## [1] doctor_str1 doctor_str2 most_important_fac n
## <0 rows> (or 0-length row.names)B. EGD
cscopy_with_egd %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.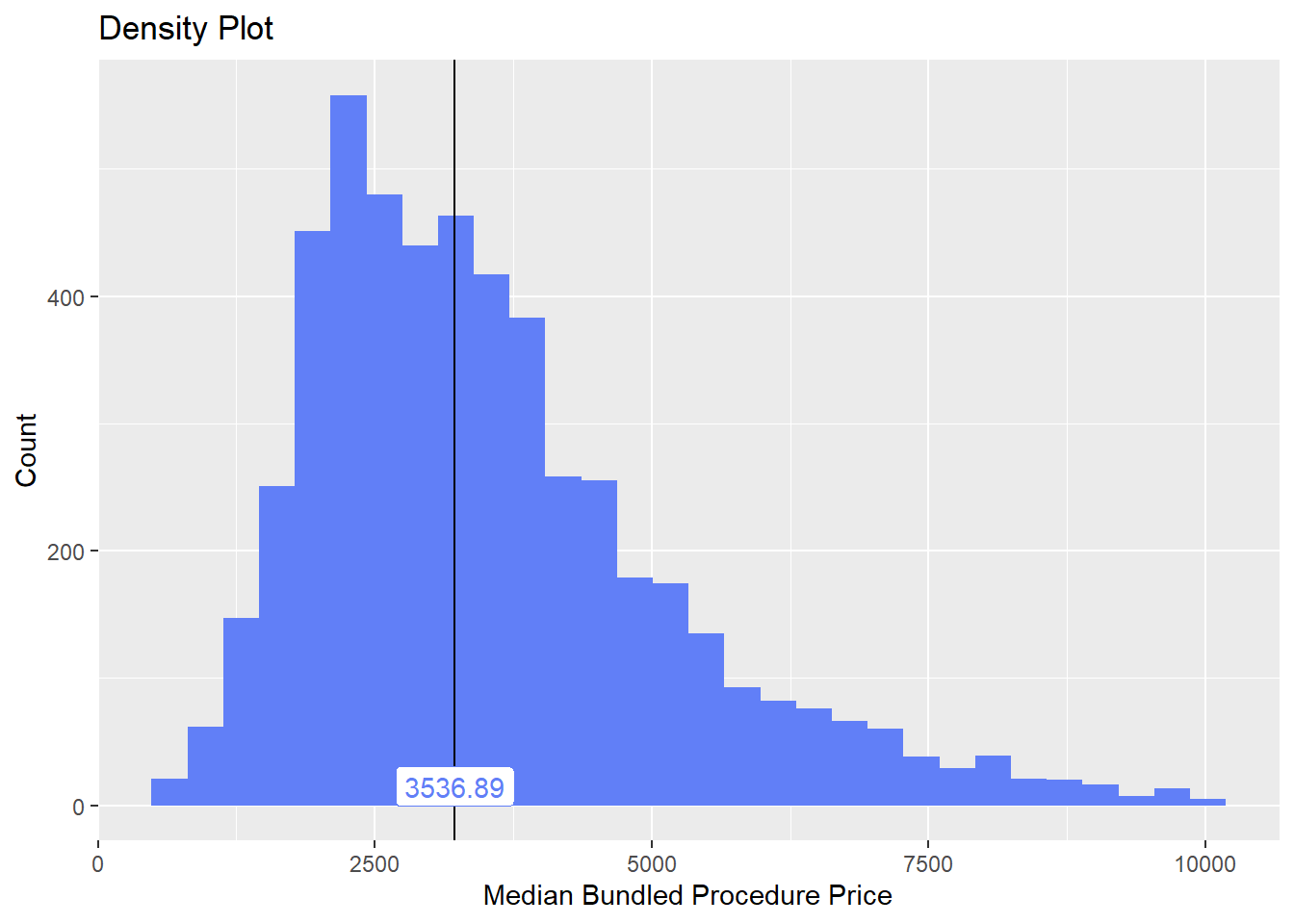
cscopy_with_egd %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 355 x 2
## name correlation
## <chr> <dbl>
## 1 medi_bun_t_sodium 0.306
## 2 surg_bun_t_small 0.27
## 3 surg_bun_t_bowel 0.270
## 4 surg_bun_t_endoscopy 0.270
## 5 surg_bun_t_endo 0.255
## 6 medi_bun_t_inject 0.226
## 7 surg_bun_t_diagnostic 0.224
## 8 anes_bun_t_anes 0.147
## 9 path_bun_t_test 0.142
## 10 path_bun_t_patho 0.139
## # ... with 345 more rowscscopy_with_egd %>% get_tag_density_information("medi_bun_t_sodium") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_sodium"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 295## $dist_plots
##
## $stat_tables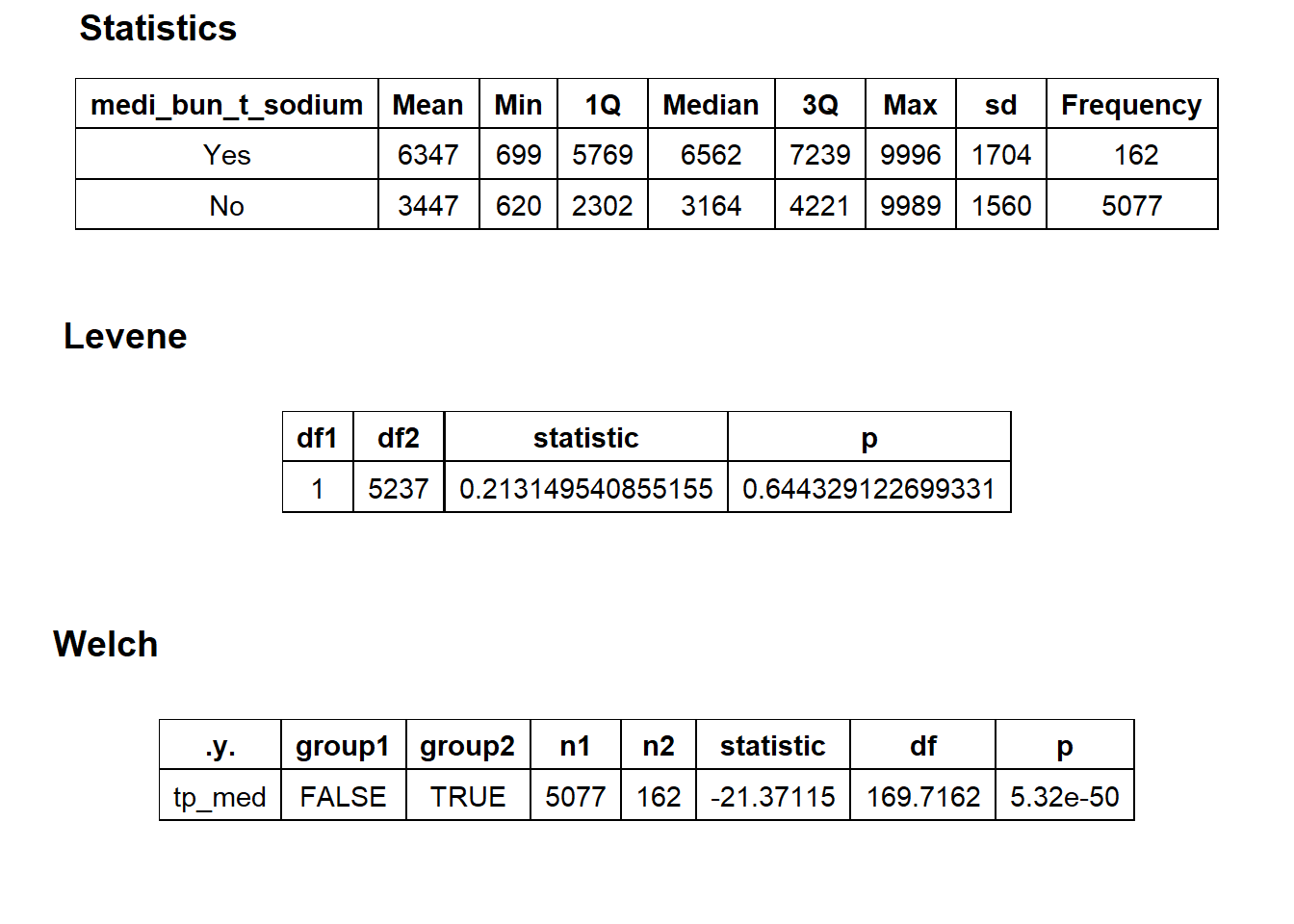
cscopy_with_edg_with_sodium <- cscopy_with_egd[medi_bun_t_sodium==T]
cscopy_with_egd <- cscopy_with_egd[medi_bun_t_sodium==F]cscopy_with_egd %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 342 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_diagnostic 0.207
## 2 medi_bun_t_inject 0.175
## 3 anes_bun_t_mod_sed 0.158
## 4 surg_bun_t_remov 0.151
## 5 surg_bun_t_removal 0.150
## 6 surg_bun_t_lesion 0.15
## 7 surg_bun_t_njx 0.141
## 8 surg_bun_t_sub 0.140
## 9 path_bun_t_patho 0.136
## 10 path_bun_t_pathologist 0.136
## # ... with 332 more rowsC. Lesion Removal
We will now break down the Colonoscopy with Lesion Removal Category down into appropriate categories.
lets check the dist and correlated tags
cscopy_with_lesion %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
cscopy_with_lesion %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 323 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_sub 0.341
## 2 surg_bun_t_njx 0.337
## 3 anes_bun_t_mod_sed 0.301
## 4 surg_bun_t_biopsy 0.297
## 5 medi_bun_t_inject 0.236
## 6 surg_bun_t_resect 0.178
## 7 anes_bun_t_anesth 0.107
## 8 faci_bun_t_dept 0.105
## 9 faci_bun_t_emergency 0.105
## 10 anes_bun_t_surg 0.105
## # ... with 313 more rowscscopy_with_lesion %>% get_tag_density_information("medi_bun_t_inject") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_inject"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 216## $dist_plots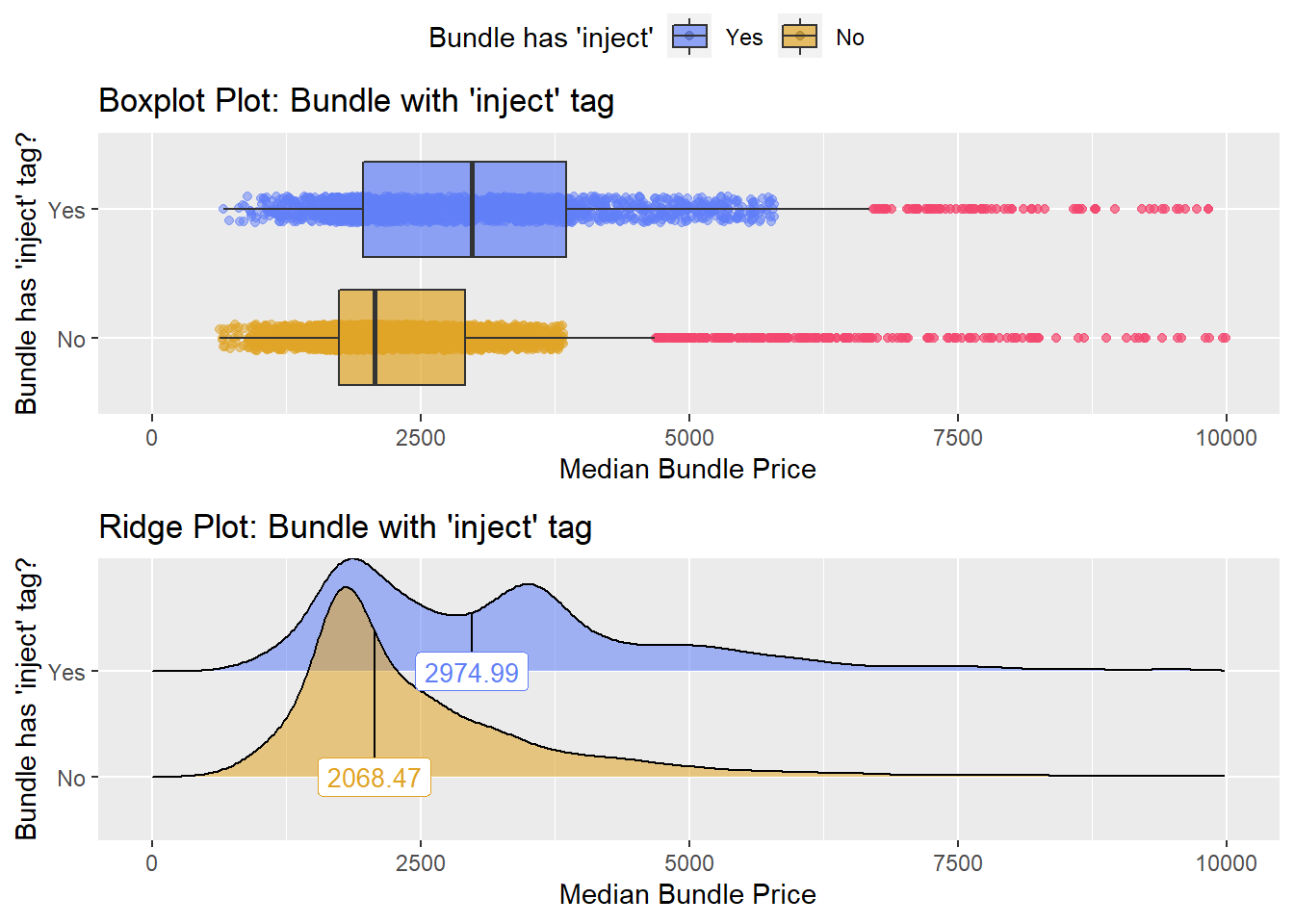
##
## $stat_tables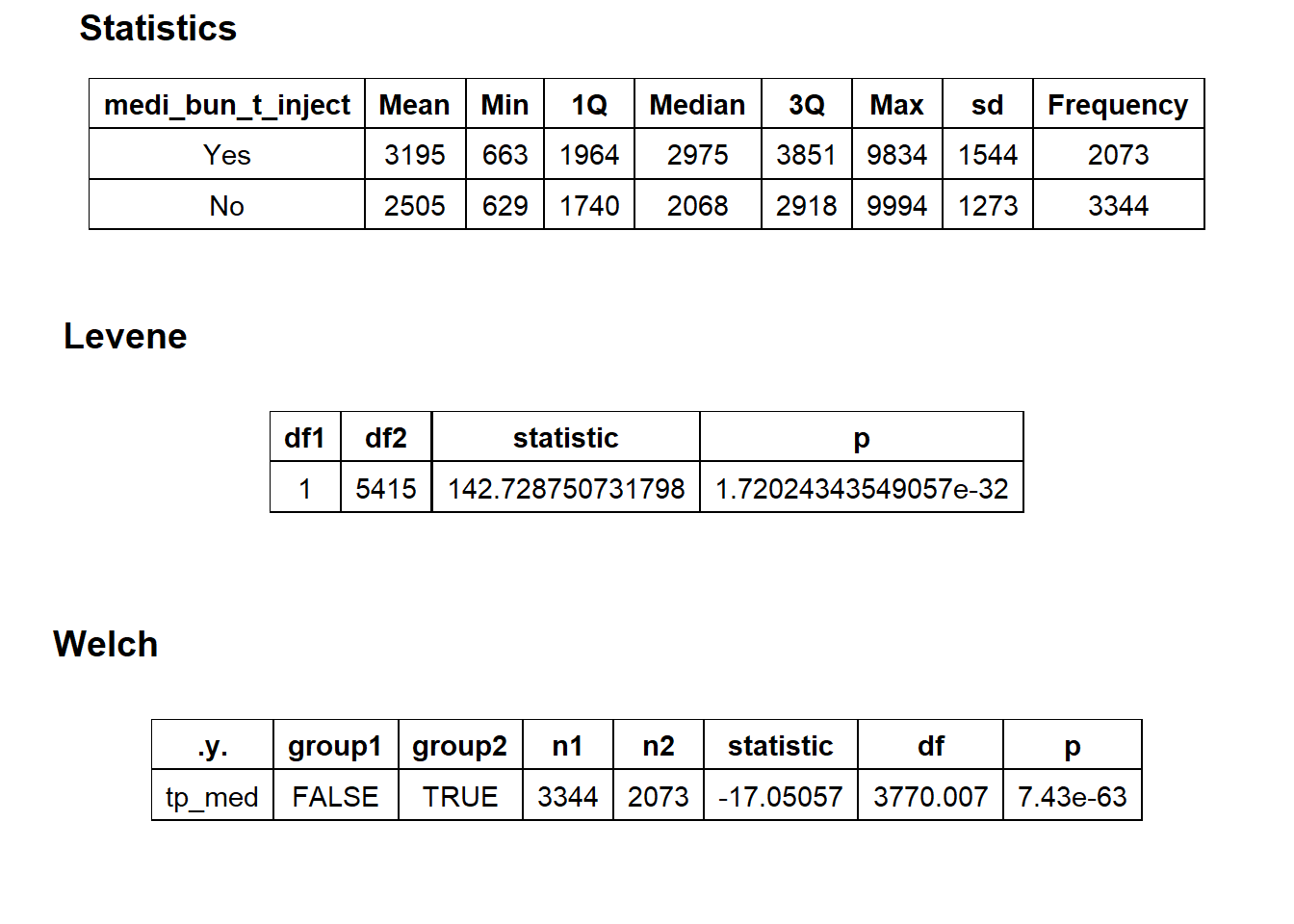 we have strong evidence to create a new group for cscopy with a lesion removal with an injection.
we have strong evidence to create a new group for cscopy with a lesion removal with an injection.
cscopy_with_lesion_with_injection <- cscopy_with_lesion[medi_bun_t_inject==T]
cscopy_with_lesion <- cscopy_with_lesion[medi_bun_t_inject==F]lets check the dist and correlated tags
cscopy_with_lesion %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.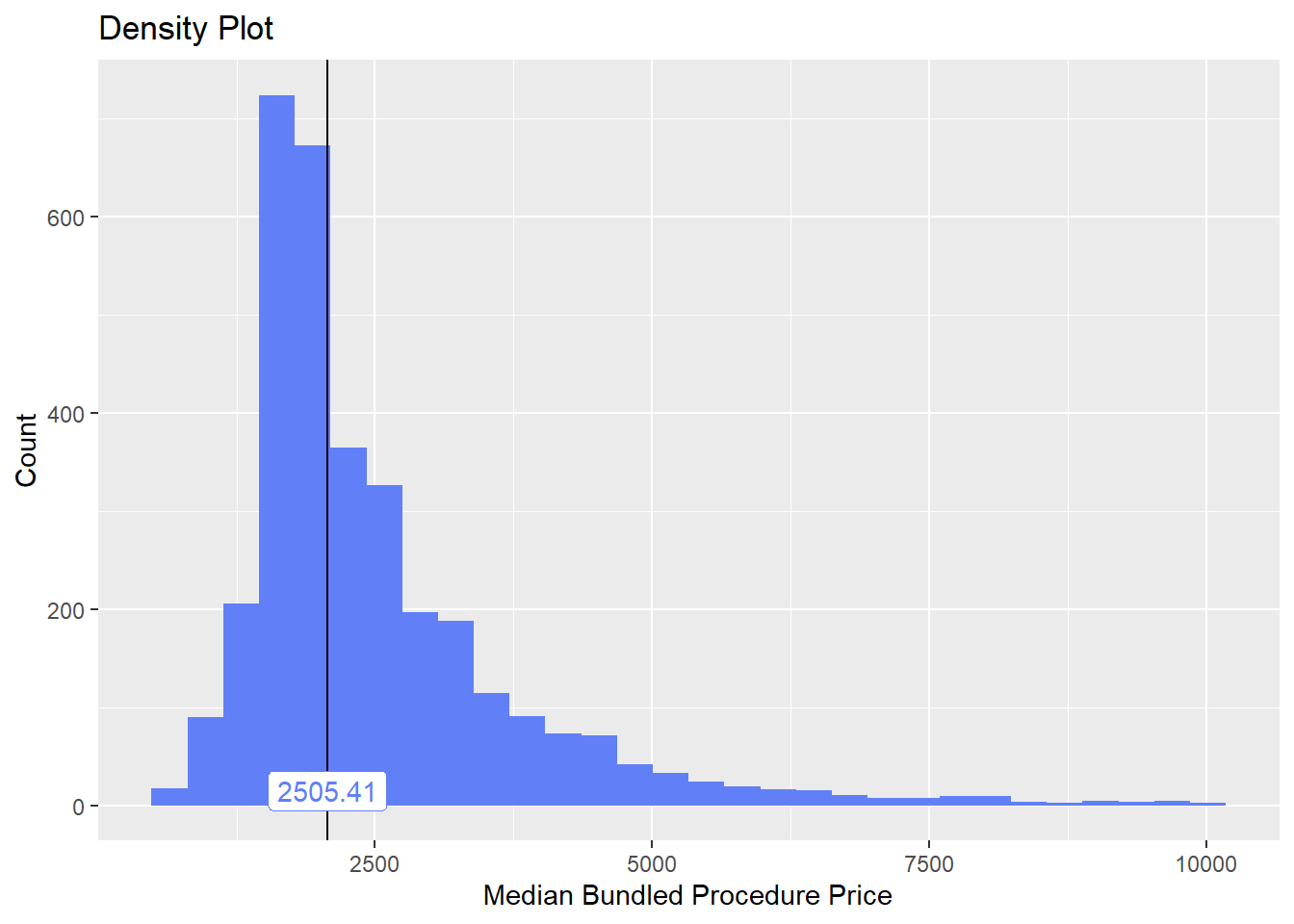
cscopy_with_lesion %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 275 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_sub 0.256
## 2 surg_bun_t_njx 0.254
## 3 surg_bun_t_biopsy 0.254
## 4 anes_bun_t_anes 0.145
## 5 faci_bun_t_dept 0.128
## 6 faci_bun_t_emergency 0.128
## 7 faci_bun_t_hospital 0.116
## 8 anes_bun_t_mod_sed 0.115
## 9 faci_bun_t_care 0.0962
## 10 faci_bun_t_initial 0.0962
## # ... with 265 more rowscscopy_with_lesion %>% get_tag_density_information("surg_bun_t_biopsy") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_biopsy"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 150## $dist_plots
##
## $stat_tables There is strong evidence that a lesion removal with a biopsy should be a separate procedure.
There is strong evidence that a lesion removal with a biopsy should be a separate procedure.
cscopy_with_lesion_and_biopsy <- cscopy_with_lesion[surg_bun_t_biopsy==T]
cscopy_with_lesion <- cscopy_with_lesion[surg_bun_t_biopsy==F]lets check the dist and correlated tags
cscopy_with_lesion %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.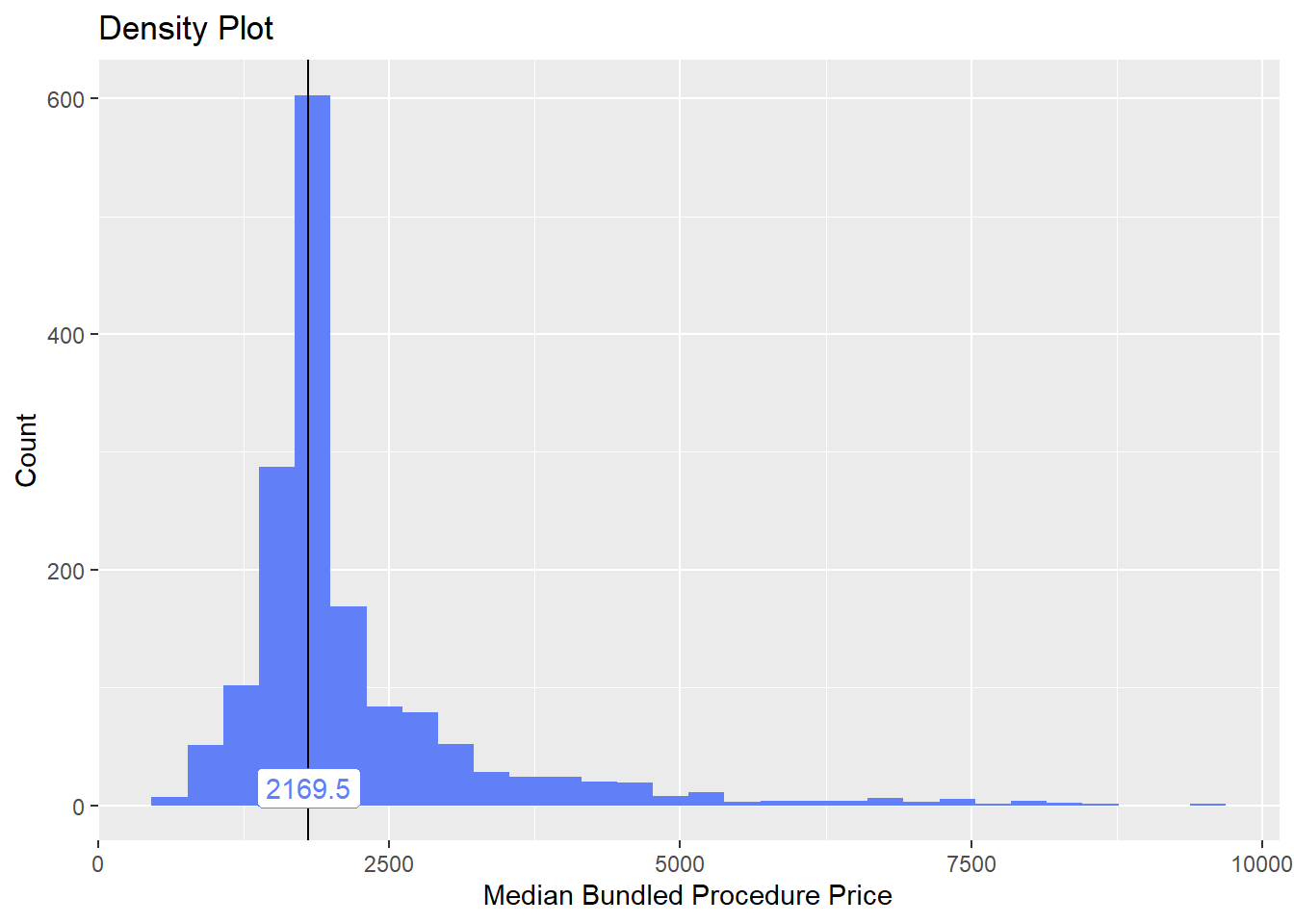
cscopy_with_lesion %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 231 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_sub 0.269
## 2 surg_bun_t_njx 0.263
## 3 faci_bun_t_dept 0.204
## 4 faci_bun_t_emergency 0.204
## 5 anes_bun_t_anes 0.180
## 6 faci_bun_t_hospital 0.168
## 7 anes_bun_t_anesth 0.139
## 8 surg_bun_t_sheath 0.134
## 9 surg_bun_t_tendon 0.134
## 10 surg_bun_t_endo 0.131
## # ... with 221 more rowsthe njx tag is picking up is “Egd us transmural injxn/mark” or “Colonoscopy submucous njx”
cscopy_with_lesion %>% get_tag_density_information("surg_bun_t_sub") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_sub"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 408## $dist_plots
##
## $stat_tables while there is a legitimate difference, it is not a large enough sample to warrant a separate procedure options
while there is a legitimate difference, it is not a large enough sample to warrant a separate procedure options
cscopy_with_lesion <- cscopy_with_lesion[surg_bun_t_sub==F]lets check the dist and correlated tags
cscopy_with_lesion %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.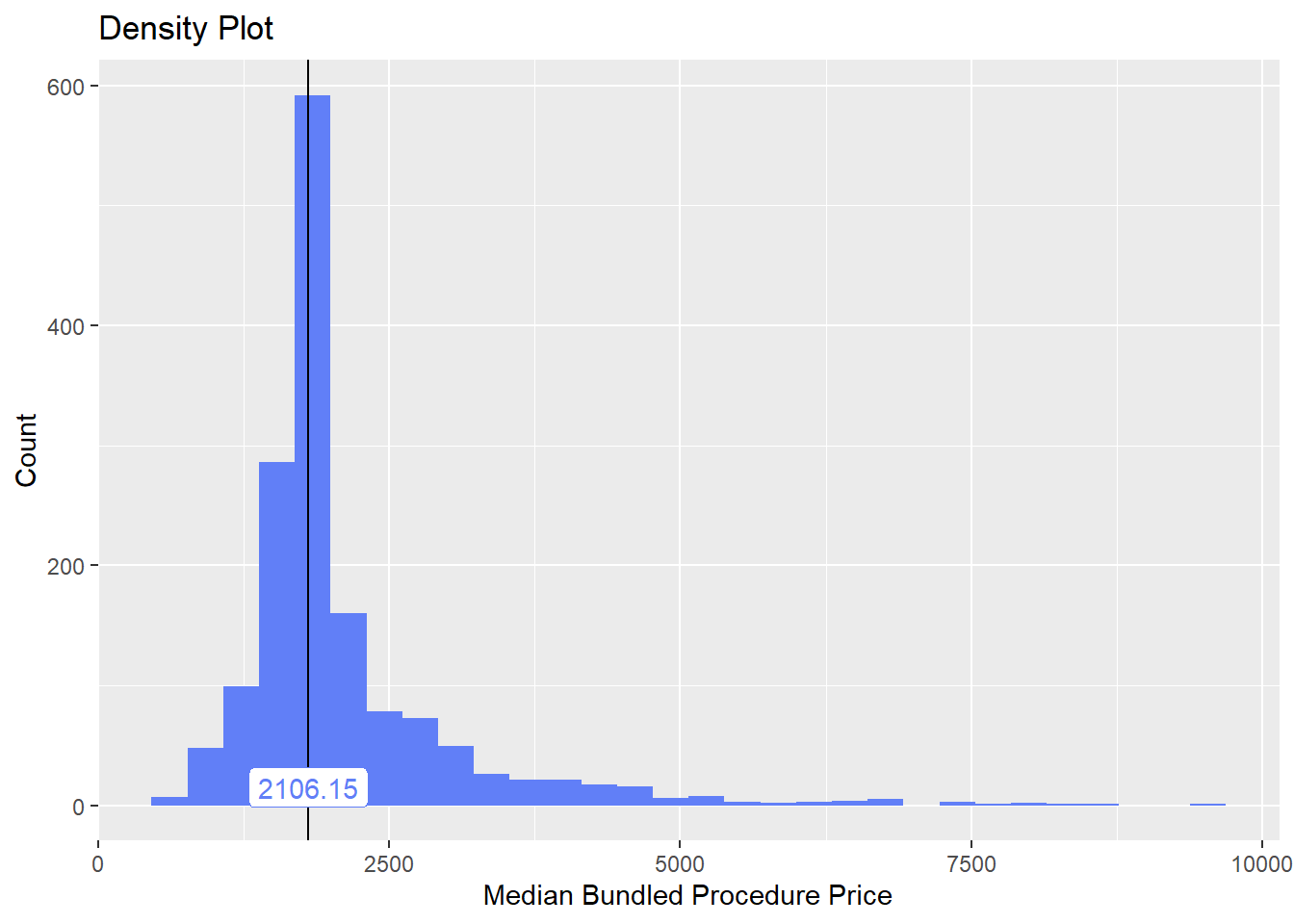
cscopy_with_lesion %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 222 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_anes 0.211
## 2 faci_bun_t_dept 0.198
## 3 faci_bun_t_emergency 0.198
## 4 surg_bun_t_sheath 0.153
## 5 surg_bun_t_tendon 0.153
## 6 surg_bun_t_endo 0.150
## 7 surg_bun_t_block 0.148
## 8 medi_bun_t_add-on 0.144
## 9 medi_bun_t_hydrate 0.144
## 10 medi_bun_t_observ 0.144
## # ... with 212 more rowscscopy_with_lesion %>% get_tag_density_information("faci_bun_t_emergency") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ faci_bun_t_emergency"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 518## $dist_plots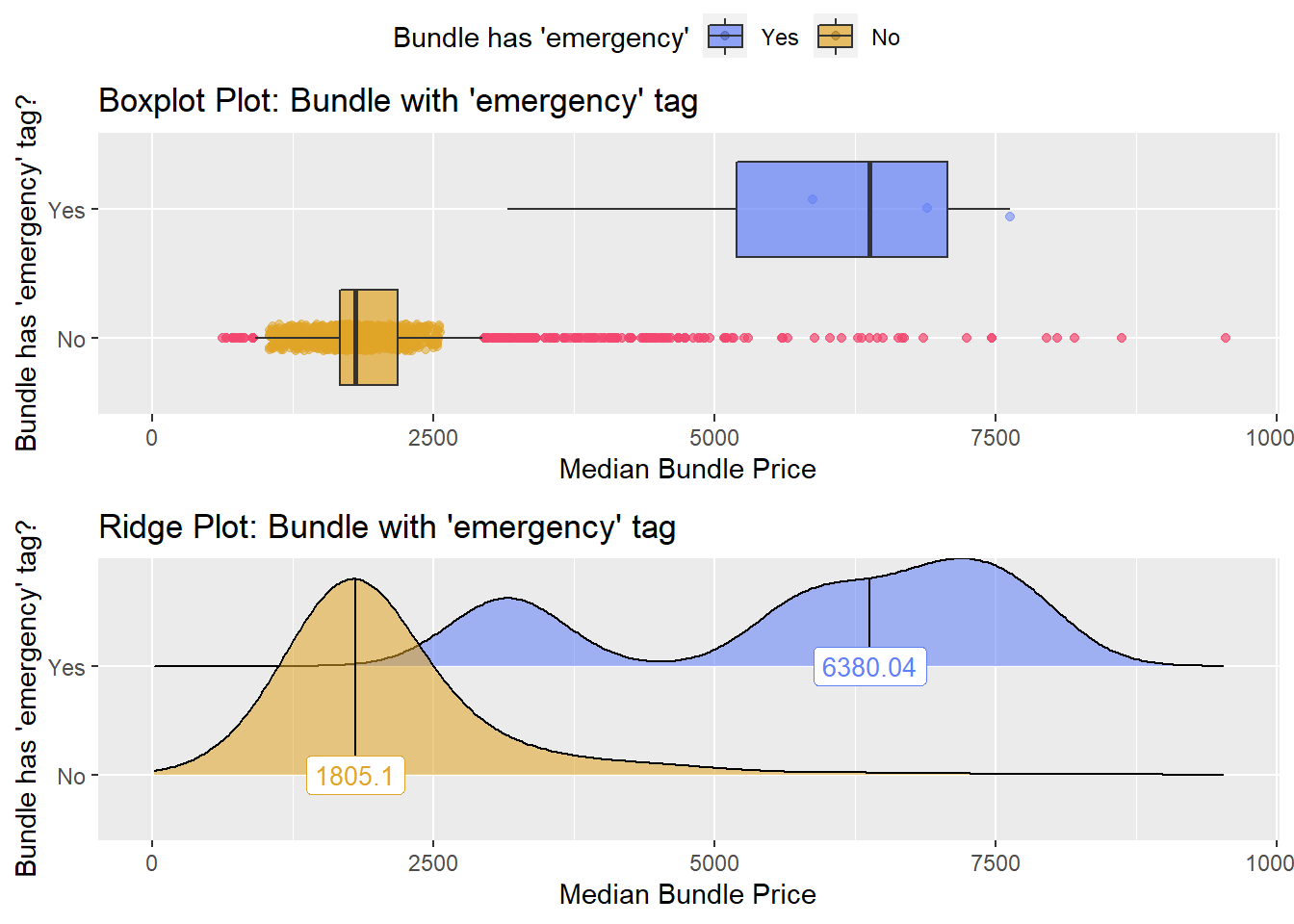
##
## $stat_tables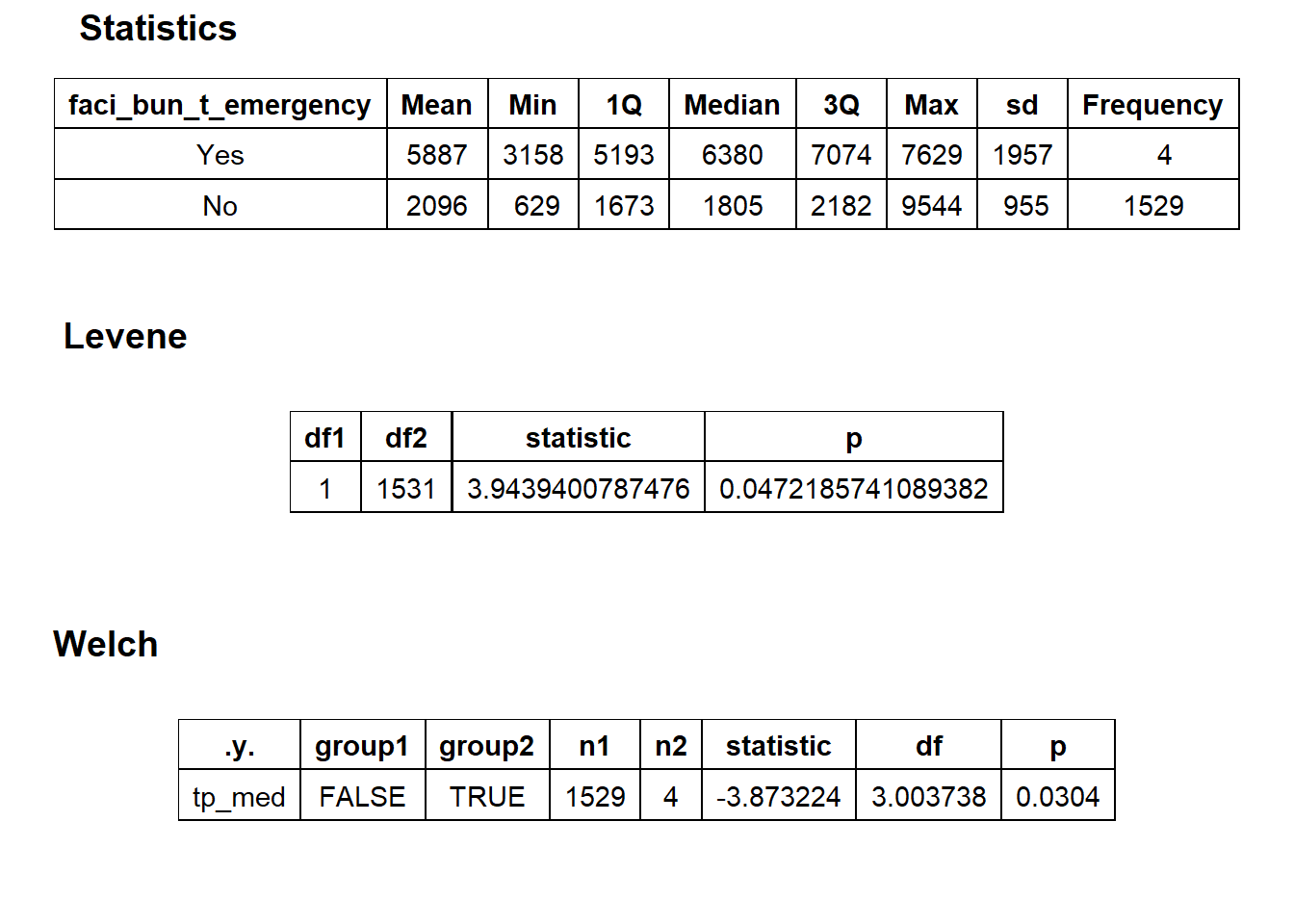 not a large enough sample to warrant a split.
not a large enough sample to warrant a split.
cscopy_with_lesion <- cscopy_with_lesion[faci_bun_t_emergency==F]lets check the dist and correlated tags
cscopy_with_lesion %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
cscopy_with_lesion %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 205 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_anes 0.206
## 2 surg_bun_t_sheath 0.157
## 3 surg_bun_t_tendon 0.157
## 4 surg_bun_t_endo 0.154
## 5 surg_bun_t_block 0.152
## 6 anes_bun_t_anesth 0.131
## 7 anes_bun_t_mod_sed 0.129
## 8 surg_bun_t_eye 0.128
## 9 radi_bun_t_guide 0.116
## 10 surg_bun_t_remove 0.115
## # ... with 195 more rowscscopy_with_lesion <- cscopy_with_lesion[surg_bun_t_sheath==F & surg_bun_t_block==F & surg_bun_t_eye==F & surg_bun_t_cystoscopy==F]cscopy_with_lesion %>% get_tag_density_information("path_bun_t_patho") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_patho"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 80.1## $dist_plots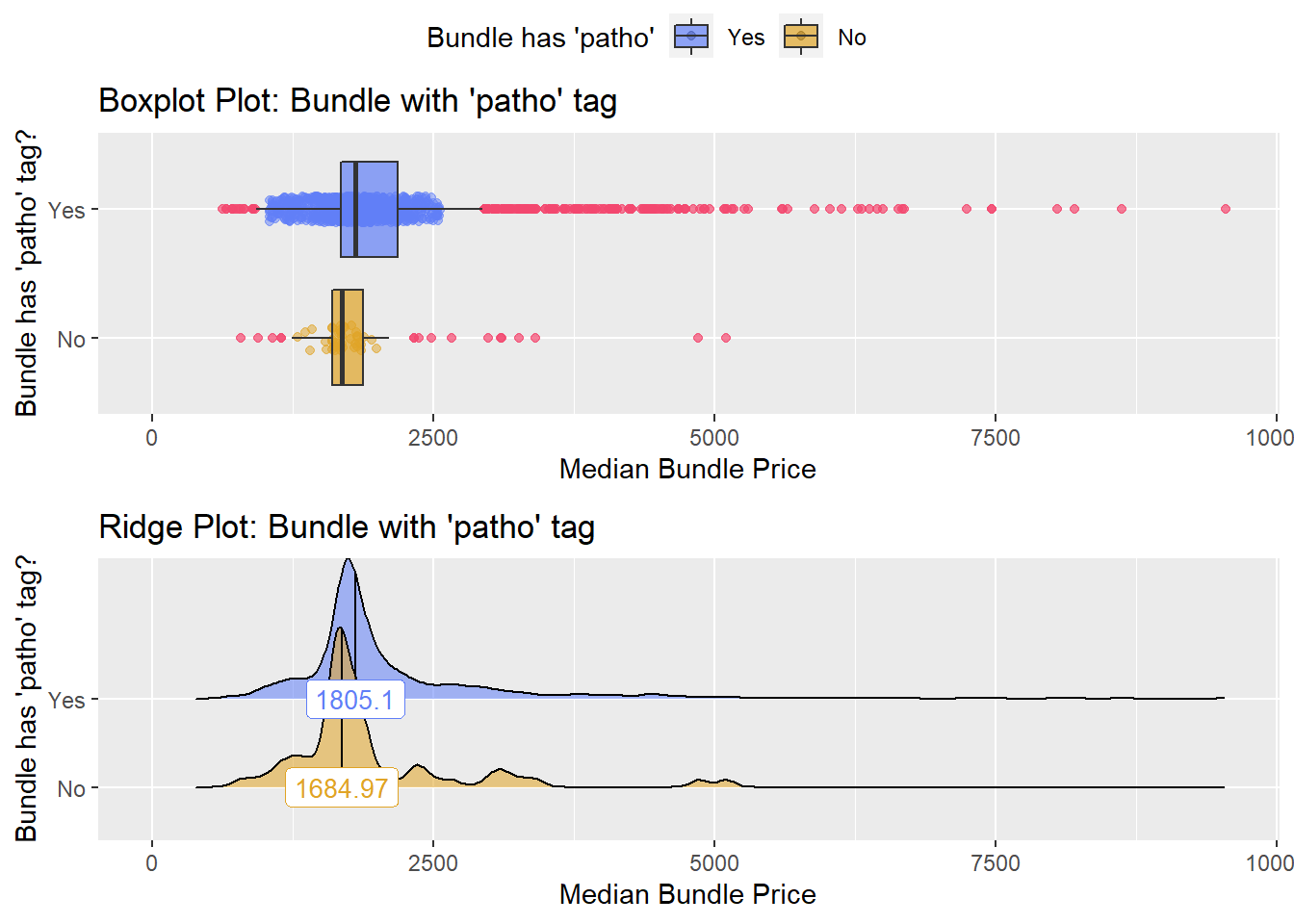
##
## $stat_tables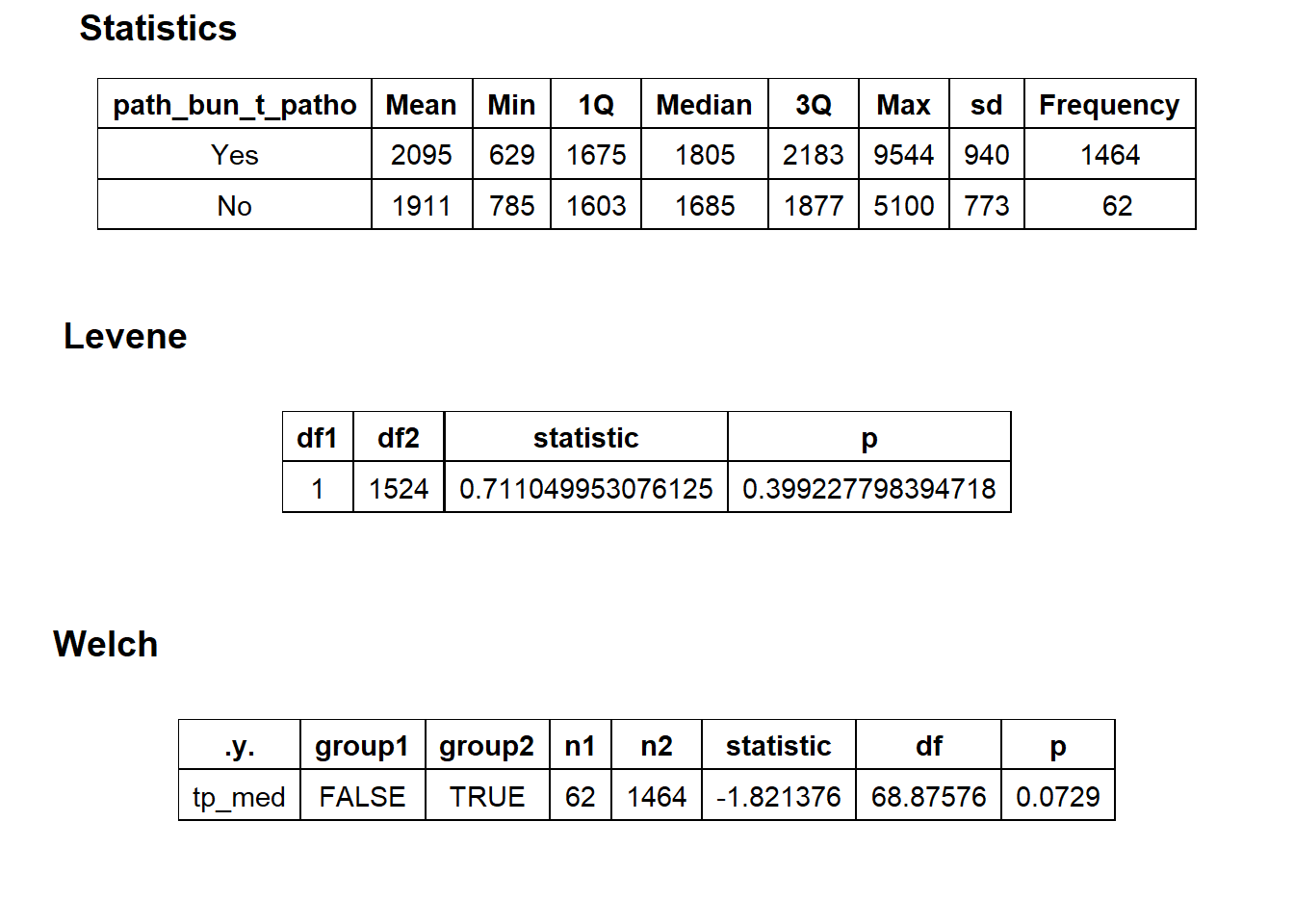 Not justified to split
Not justified to split
lets check the dist and correlated tags
cscopy_with_lesion %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.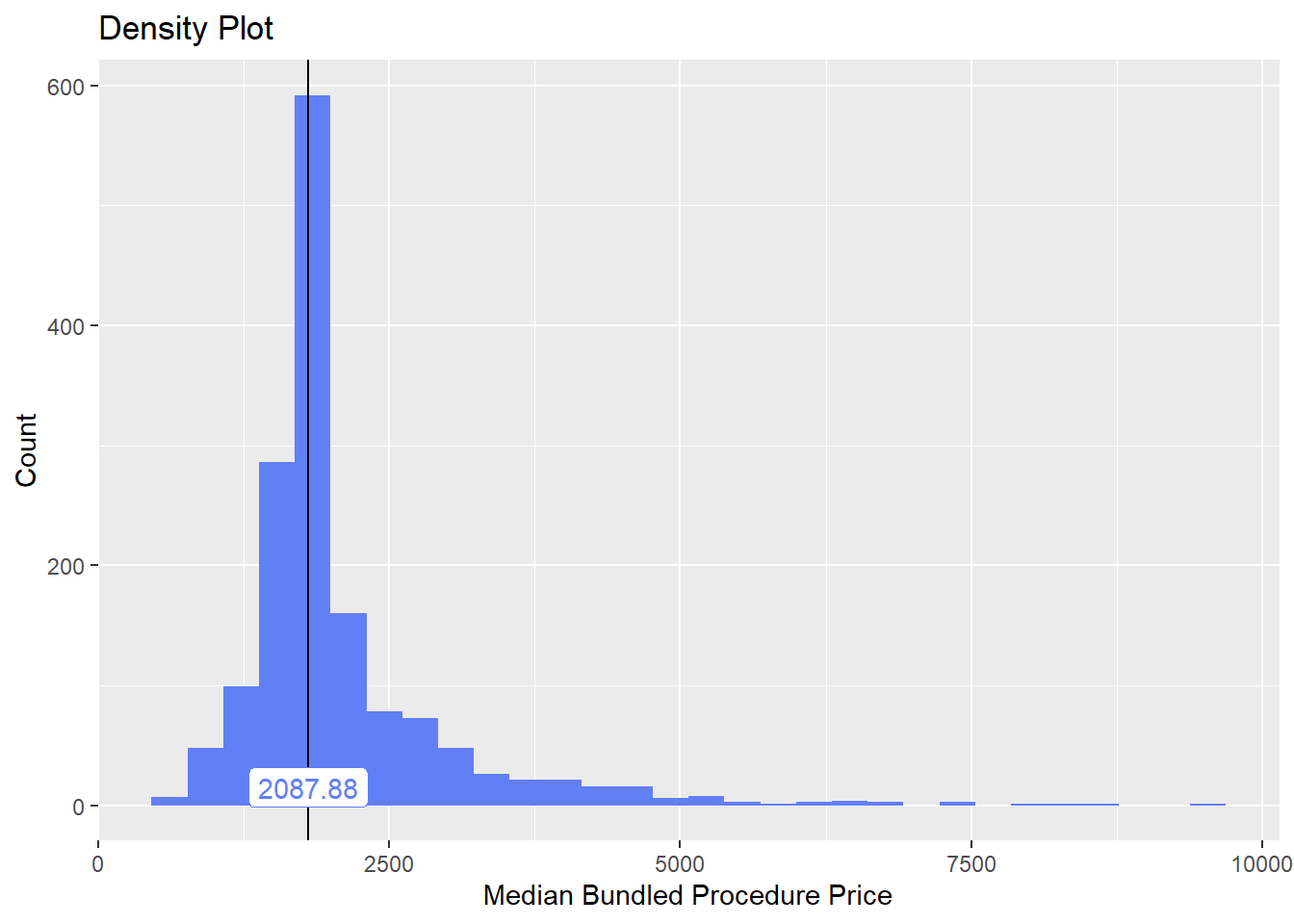
cscopy_with_lesion %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 198 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_anes 0.182
## 2 anes_bun_t_mod_sed 0.130
## 3 medi_bun_t_normal 0.105
## 4 medi_bun_t_saline 0.105
## 5 medi_bun_t_solution 0.105
## 6 path_bun_t_agent 0.0698
## 7 path_bun_t_reagent 0.0698
## 8 surg_bun_t_endo 0.0628
## 9 surg_bun_t_esoph 0.0628
## 10 surg_bun_t_exam 0.0628
## # ... with 188 more rowscscopy_with_lesion_btbv4 <- cscopy_with_lesion %>% btbv4()cscopy_with_lesion_btbv4 <- cscopy_with_lesion_btbv4 %>%
filter(tp_cnt_cnt>5) %>%
select(doctor_str1,
doctor_str2,
doctor_npi_str1,
doctor_npi_str2,
most_important_fac,
most_important_fac_npi,
tp_med_med,
tp_med_surg,
tp_med_medi,
tp_med_path,
tp_med_radi,
tp_med_anes,
tp_med_faci,
tp_cnt_cnt) %>%
mutate(procedure_type = 1,
procedure_modifier = "Lesion Removal")
cscopy_with_lesion_btbv4## doctor_str1 doctor_str2 doctor_npi_str1
## 1: KURT O BODILY <NA> 1780600312
## 2: JOHN C CAPENER <NA> 1750355624
## 3: CASEY RAY OWENS <NA> 1437348943
## 4: VIKRAM GARG <NA> 1760491054
## 5: ALLEN CLARK GUNNERSON <NA> 1407081524
## ---
## 226: STEVEN GREGG DESAUTELS <NA> 1790891430
## 227: C DAVID HANSEN MATTHEW EDWARD FEURER 1548366743
## 228: NOT SPECIFIED <NA> 0
## 229: DAVID J FRANTZ MATTHEW G. OLLERTON 1700092004
## 230: KURT O BODILY TROY LUNCEFORD 1780600312
## doctor_npi_str2 most_important_fac
## 1: <NA> CENTRAL UTAH SURGICAL CENTER (PROVO)
## 2: <NA> IHC HEALTH SERVICES (RIVERTON HOSPITAL)
## 3: <NA> CENTRAL UTAH CLINIC (REVERE HEALTH)
## 4: <NA> IHC HEALTH SERVICES (LOGAN REGIONAL HOSPITAL)
## 5: <NA> IHC HEALTH SERVICES (DIXIE REGIONAL MEDICAL CENTER)
## ---
## 226: <NA> NOT SPECIFIED
## 227: 1508157942 IHC HEALTH SERVICES (LDS HOSPITAL)
## 228: <NA> DIABETES RELIEF UTAH OGDEN (LAYTON)
## 229: 1518959733 IHC HEALTH SERVICES (AMERICAN FORK HOSPITAL)
## 230: 1194842286 IHC HEALTH SERVICES (UTAH VALLEY HOSPITAL)
## most_important_fac_npi tp_med_med tp_med_surg tp_med_medi tp_med_path
## 1: 1962463950 1153.580 1043.365 51.050 0.000
## 2: 1154551919 2002.390 1214.882 9.675 130.725
## 3: 1437205028 979.300 884.890 0.000 79.470
## 4: 1831108497 2146.510 1669.920 140.910 211.420
## 5: 1366452880 3944.450 2851.970 64.175 183.280
## ---
## 226: 0 1536.175 938.020 0.000 111.630
## 227: 1528078581 1614.750 1245.030 57.740 90.730
## 228: 1619402799 949.720 377.720 329.870 92.700
## 229: 1912014358 1677.710 1591.950 0.000 0.000
## 230: 1114025491 2057.230 1672.360 0.000 126.860
## tp_med_radi tp_med_anes tp_med_faci tp_cnt_cnt procedure_type
## 1: 0 59.165 0.0000 13 1
## 2: 0 0.000 672.2075 350 1
## 3: 0 14.940 0.0000 31 1
## 4: 0 0.000 65.1100 1021 1
## 5: 0 0.000 1005.2200 430 1
## ---
## 226: 0 486.525 0.0000 6 1
## 227: 0 0.000 221.2500 9 1
## 228: 0 15.100 134.3300 9 1
## 229: 0 0.000 85.7600 7 1
## 230: 0 15.310 242.7000 9 1
## procedure_modifier
## 1: Lesion Removal
## 2: Lesion Removal
## 3: Lesion Removal
## 4: Lesion Removal
## 5: Lesion Removal
## ---
## 226: Lesion Removal
## 227: Lesion Removal
## 228: Lesion Removal
## 229: Lesion Removal
## 230: Lesion Removal1. injection
lets break down the lesion removal with an injection
cscopy_with_lesion_with_injection %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 236 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_sub 0.415
## 2 surg_bun_t_njx 0.411
## 3 anes_bun_t_mod_sed 0.392
## 4 surg_bun_t_biopsy 0.331
## 5 surg_bun_t_resect 0.217
## 6 anes_bun_t_anesth 0.140
## 7 anes_bun_t_surg 0.140
## 8 radi_bun_t_view 0.124
## 9 surg_bun_t_remove 0.115
## 10 medi_bun_t_choline 0.108
## # ... with 226 more rowscscopy_with_lesion_with_injection %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.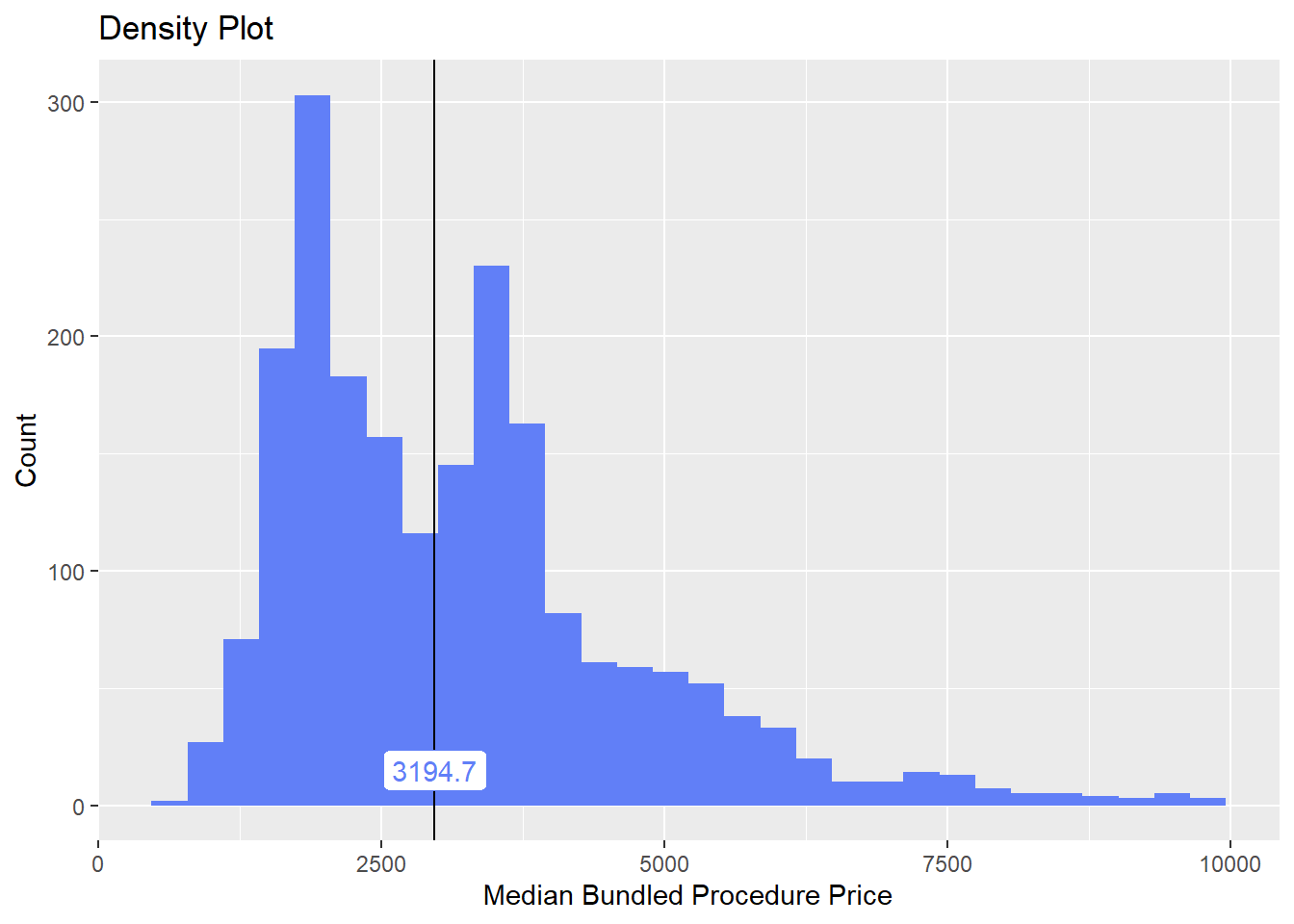
cscopy_with_lesion_with_injection %>% get_tag_density_information("surg_bun_t_sub") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_sub"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 337## $dist_plots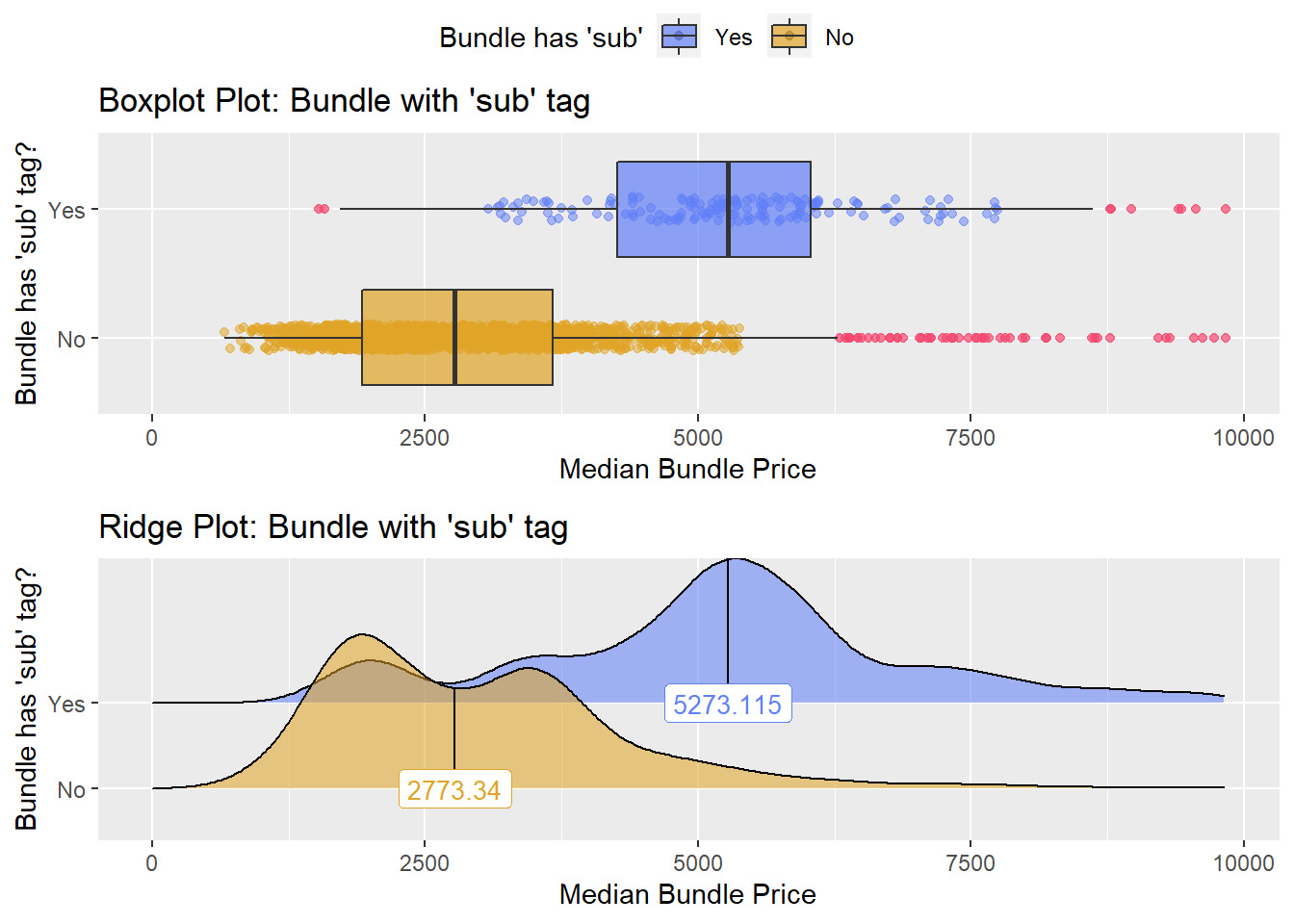
##
## $stat_tables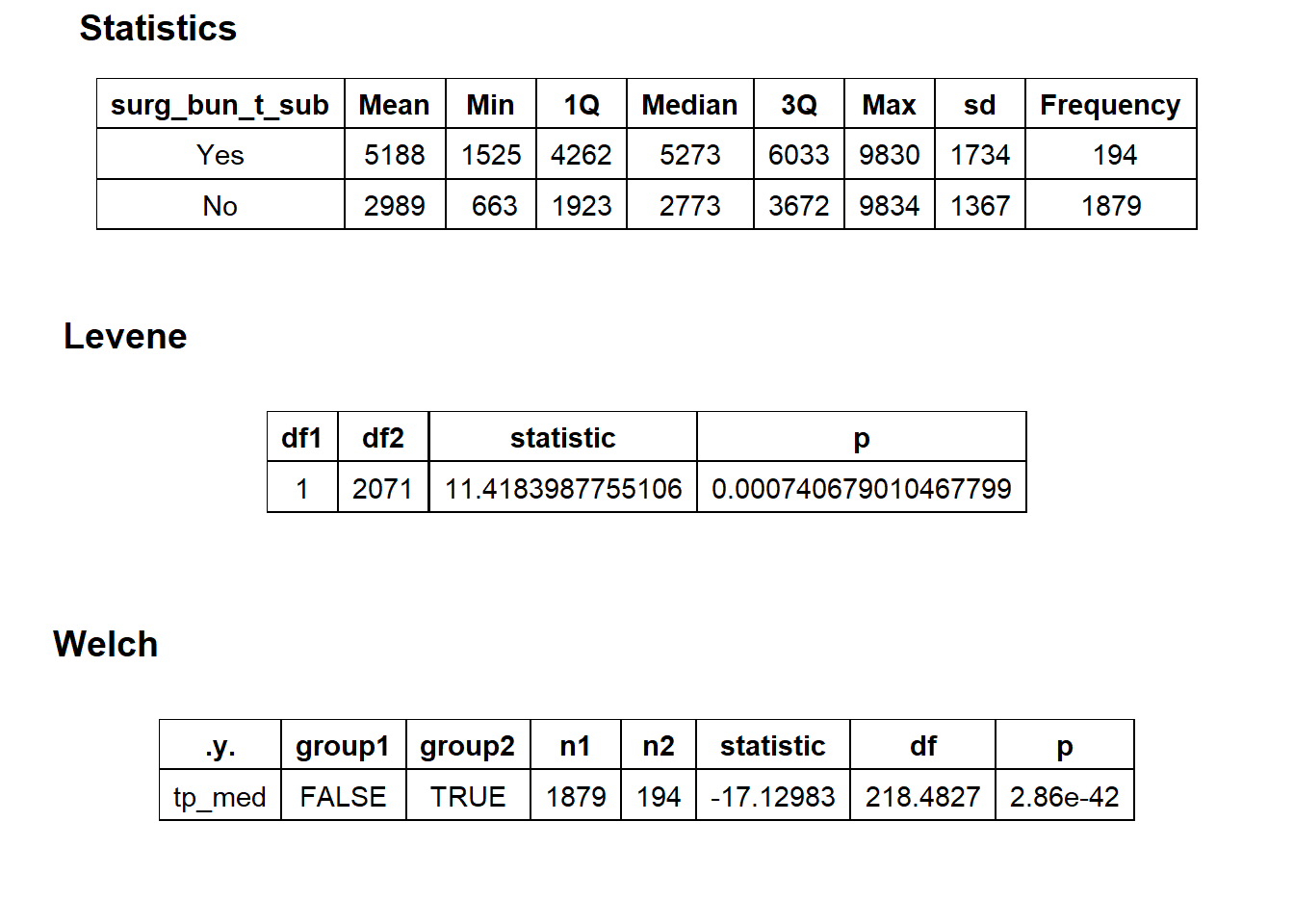
cscopy_with_lesion_with_injection <- cscopy_with_lesion_with_injection[surg_bun_t_sub==F]cscopy_with_lesion_with_injection %>% get_tag_density_information("surg_bun_t_biopsy") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_biopsy"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 207## $dist_plots
##
## $stat_tables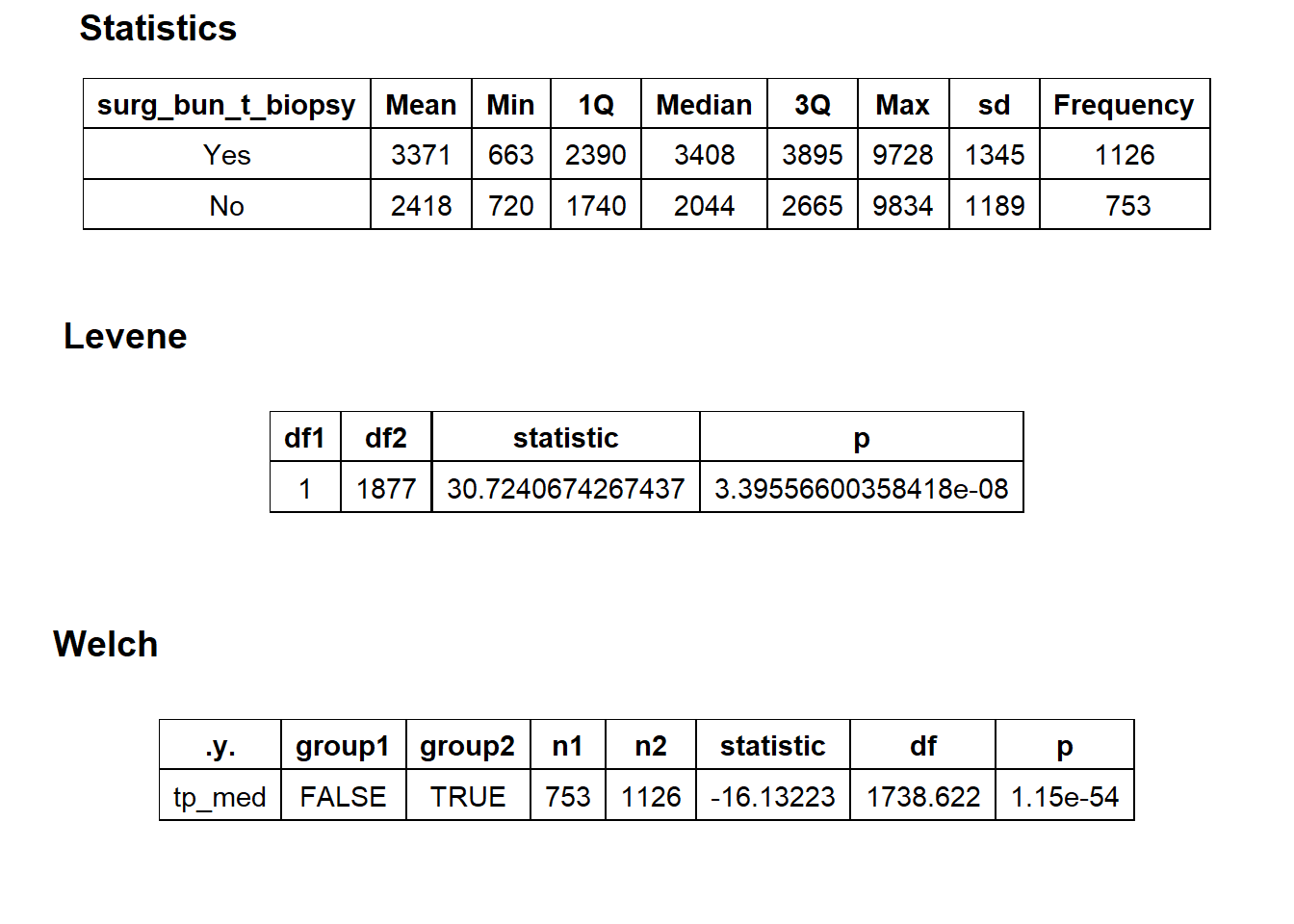
cscopy_with_lesion_biopsy_injection <- cscopy_with_lesion_with_injection[surg_bun_t_biopsy==T]
cscopy_with_lesion_with_injection <- cscopy_with_lesion_with_injection[surg_bun_t_biopsy==F]cscopy_with_lesion_with_injection %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 135 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_resect 0.308
## 2 anes_bun_t_anesth 0.256
## 3 anes_bun_t_surg 0.256
## 4 anes_bun_t_mod_sed 0.244
## 5 anes_bun_t_anes 0.236
## 6 surg_bun_t_remove 0.205
## 7 surg_bun_t_endo 0.195
## 8 surg_bun_t_tendon 0.195
## 9 surg_bun_t_wrist 0.195
## 10 anes_bun_t_lower 0.195
## # ... with 125 more rowscscopy_with_lesion_with_injection %>% get_tag_density_information("surg_bun_t_remove") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_remove"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 975## $dist_plots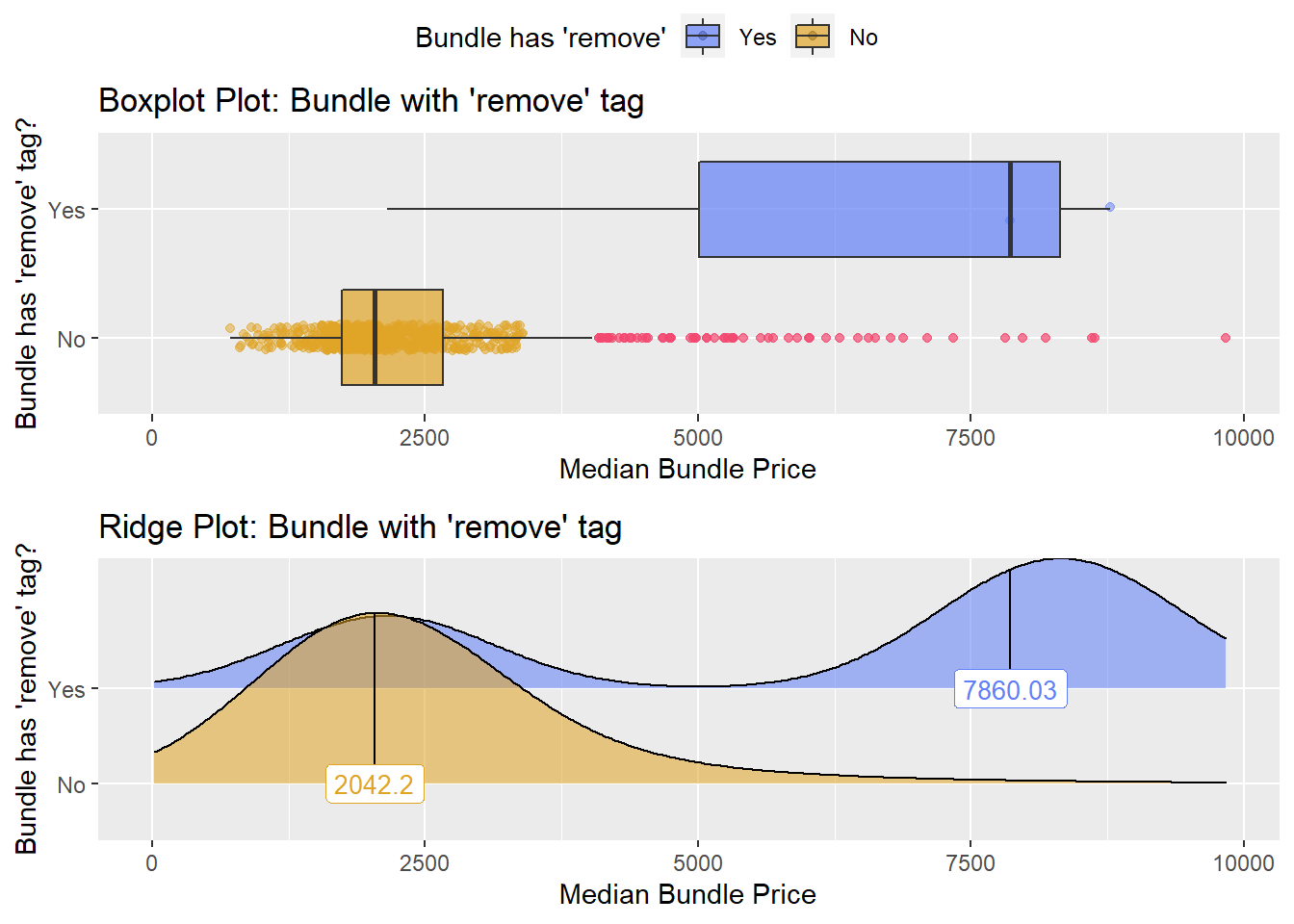
##
## $stat_tables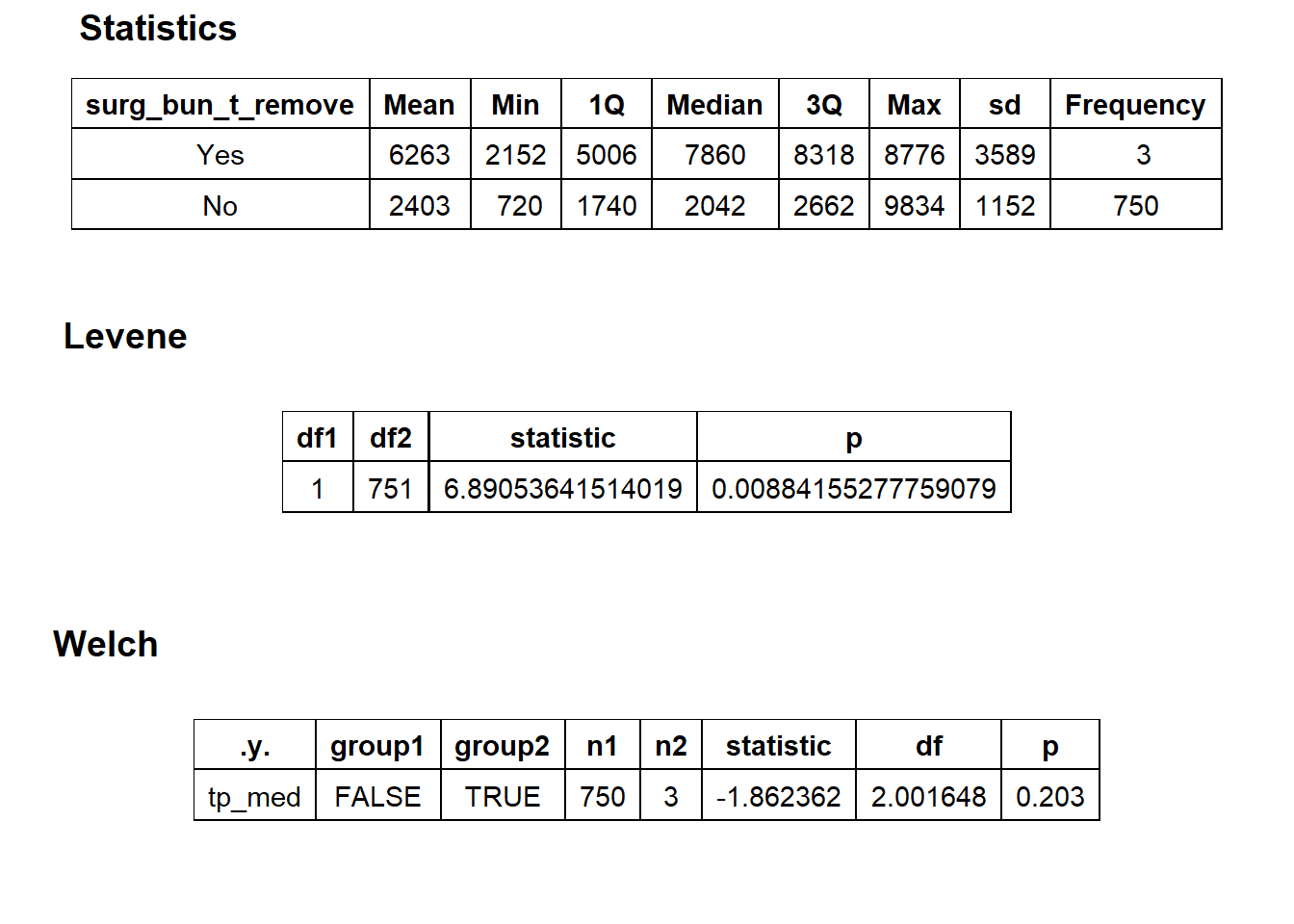
cscopy_with_lesion_with_injection <- cscopy_with_lesion_with_injection[surg_bun_t_resect==F & surg_bun_t_remove==F & medi_bun_t_normal==F ]cscopy_with_lesion_with_injection %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.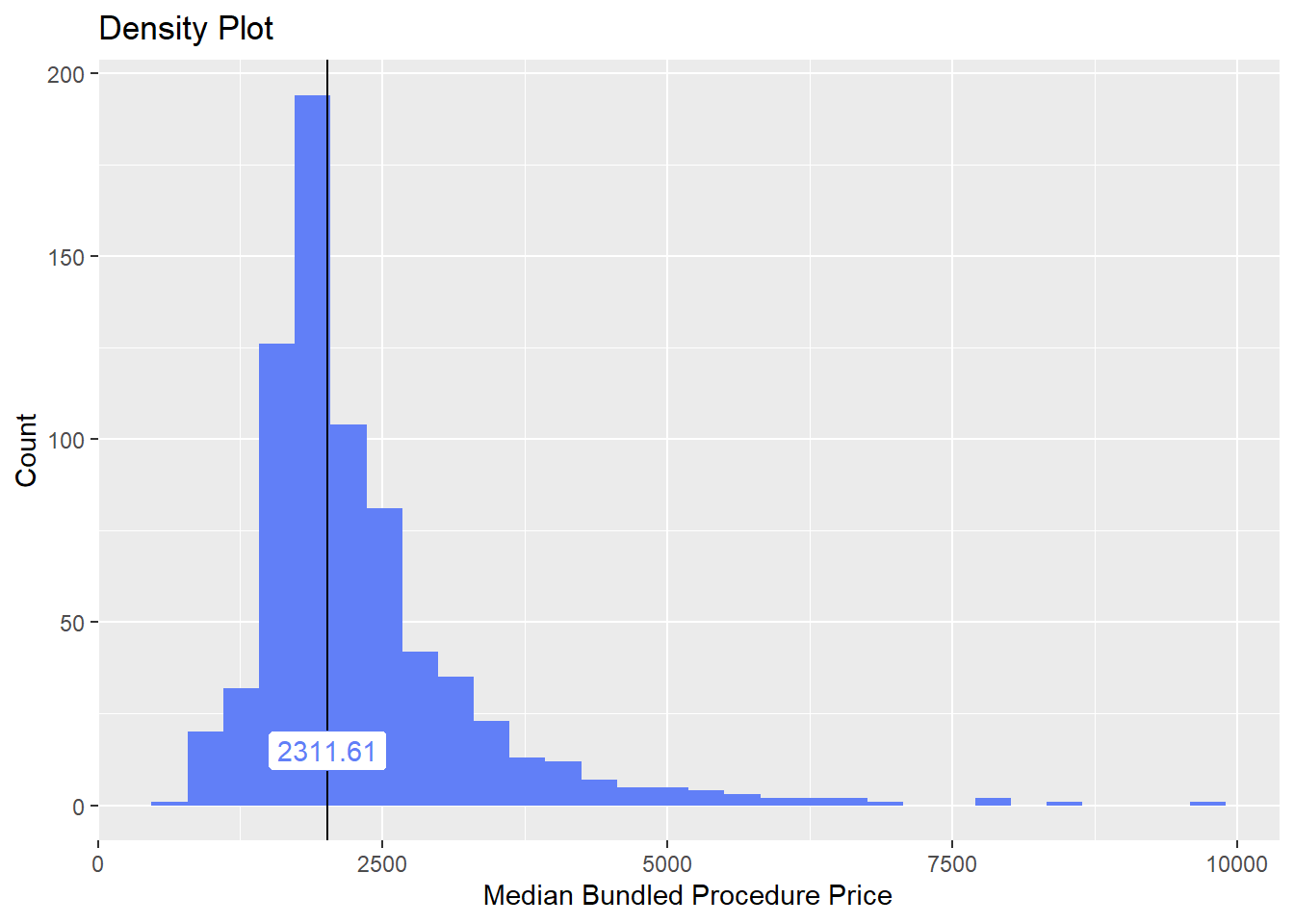
cscopy_with_lesion_with_injection_bundle <- cscopy_with_lesion_with_injection %>% btbv4()# cscopy_with_lesion_with_injection_bundle %>% View()# cscopy_with_lesion_with_injection_bundle %>% saveRDS("dash_colonoscopy_lesion_injection.RDS")2. biopsy
cscopy_with_lesion_and_biopsy %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 186 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_njx 0.245
## 2 surg_bun_t_sub 0.245
## 3 anes_bun_t_mod_sed 0.139
## 4 surg_bun_t_ligation 0.118
## 5 duration_mean 0.117
## 6 surg_bun_t_control 0.113
## 7 duration_max 0.0995
## 8 surg_bun_t_dura 0.0962
## 9 anes_bun_t_anes 0.0858
## 10 faci_bun_t_care 0.0855
## # ... with 176 more rowscscopy_with_lesion_and_biopsy %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.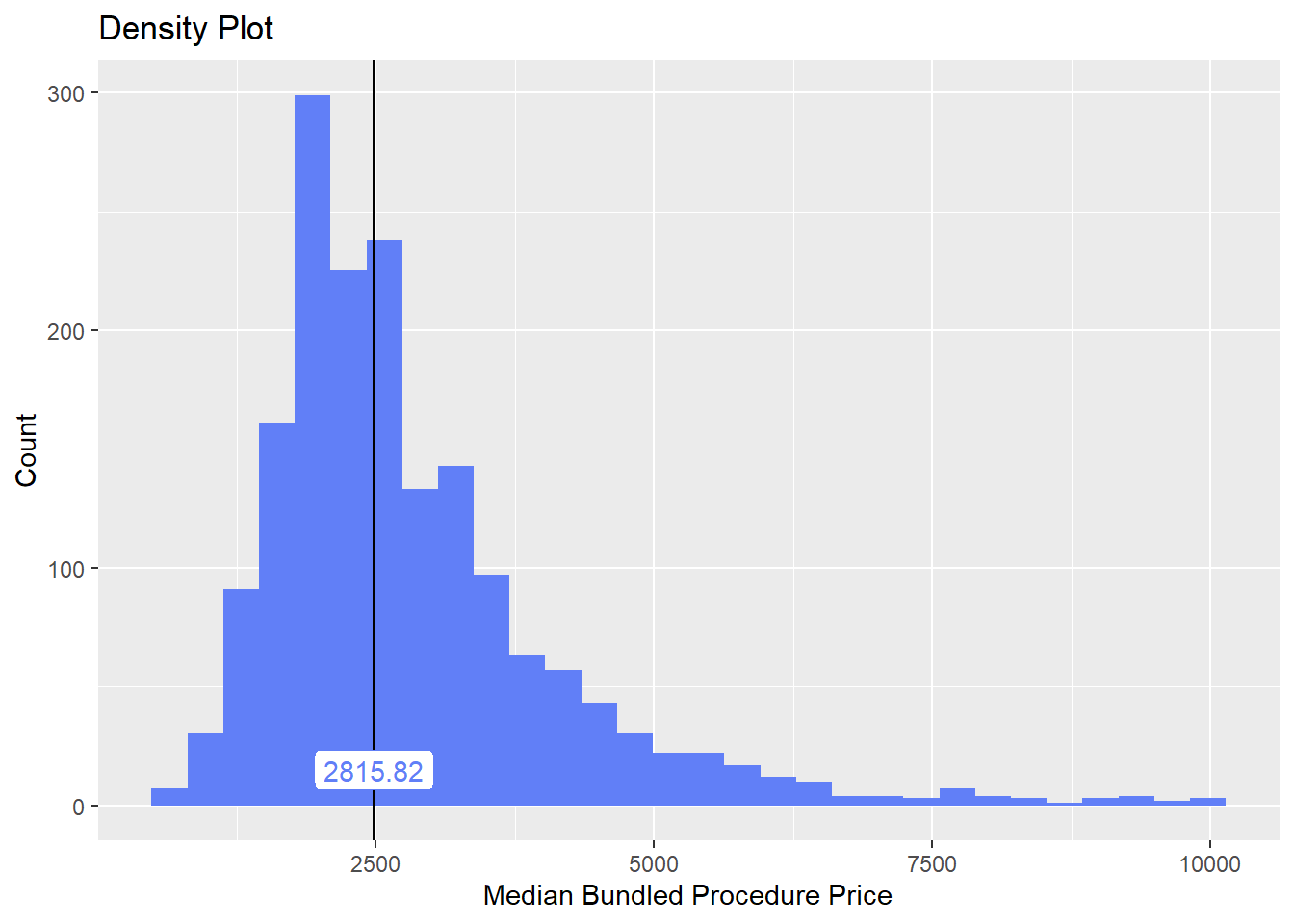
cscopy_with_lesion_and_biopsy %>% get_tag_density_information("surg_bun_t_njx") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_njx"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 407## $dist_plots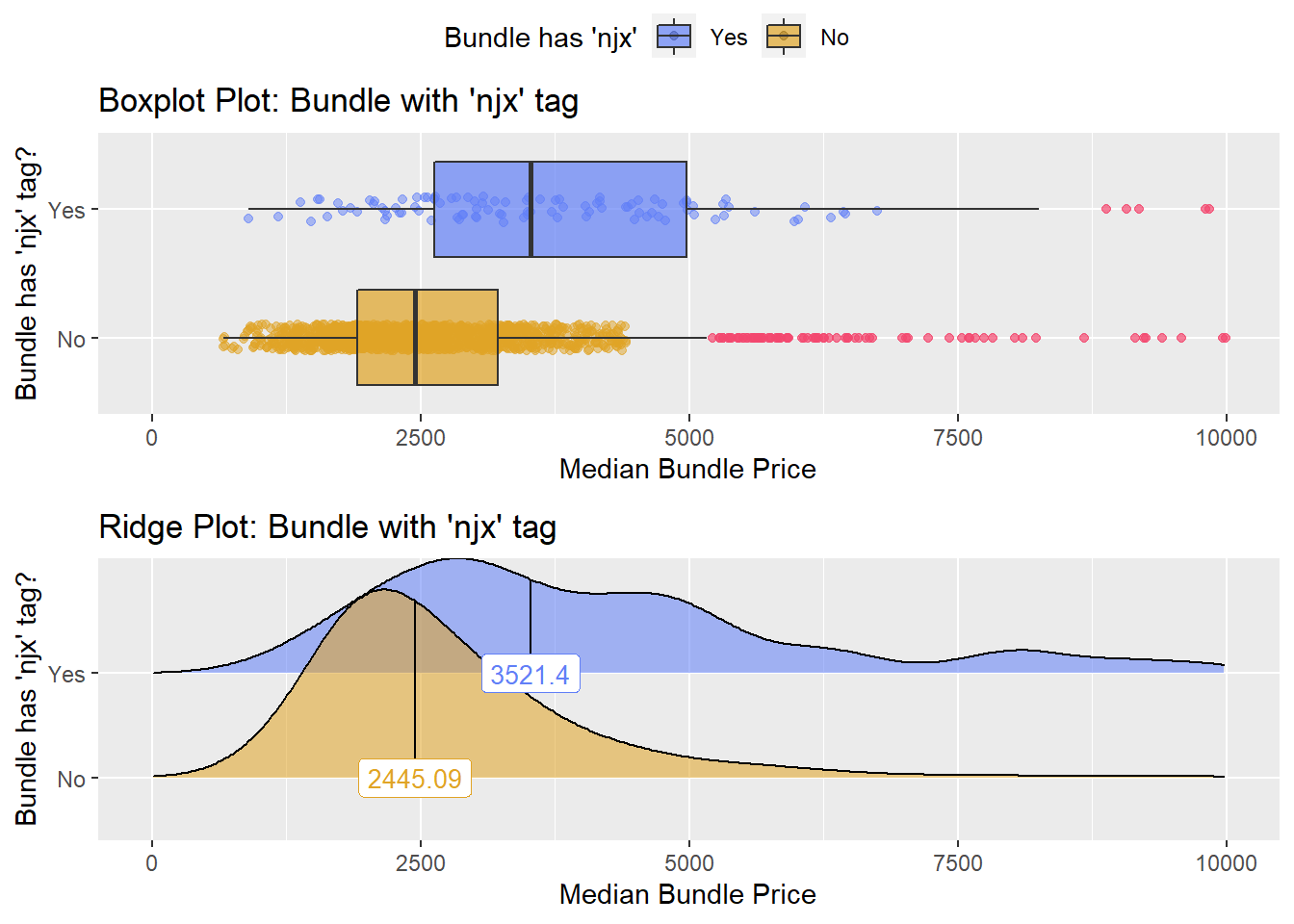
##
## $stat_tables
cscopy_with_lesion_and_biopsy <- cscopy_with_lesion_and_biopsy[surg_bun_t_njx==F & surg_bun_t_sub==F]cscopy_with_lesion_and_biopsy %>% get_tag_density_information("path_bun_t_patho") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_patho"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 338## $dist_plots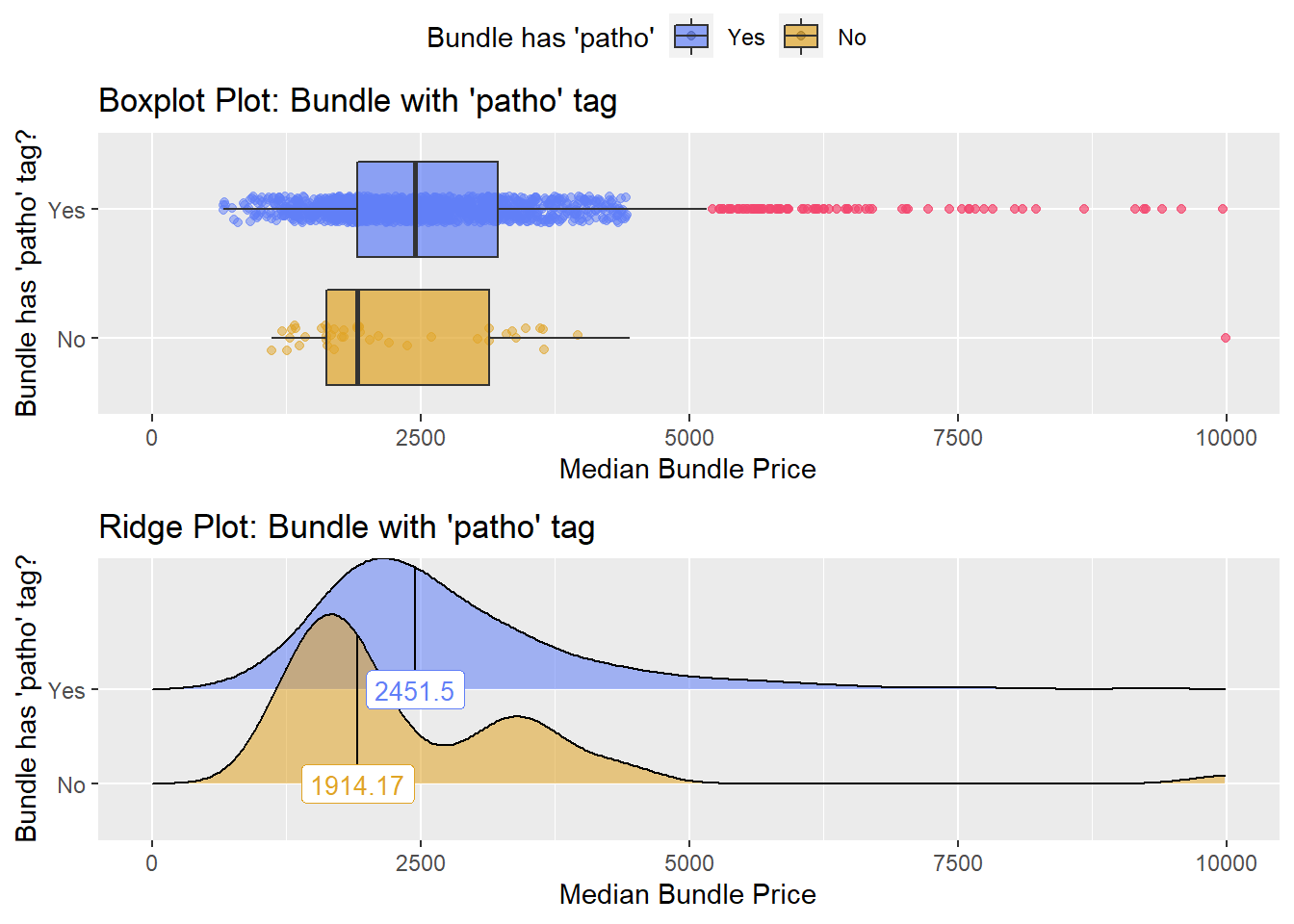
##
## $stat_tables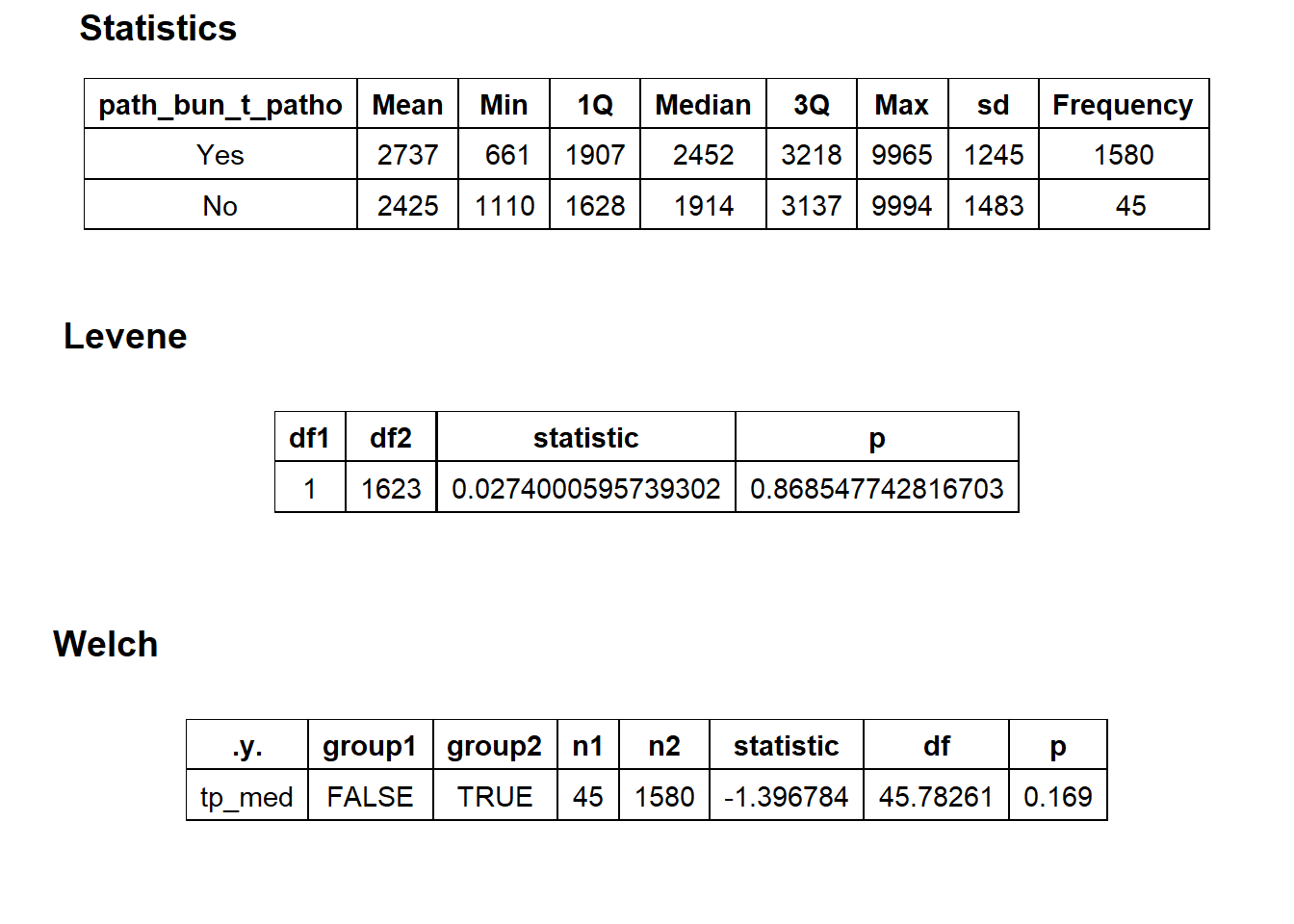
cscopy_with_lesion_and_biopsy <- cscopy_with_lesion_and_biopsy[path_bun_t_patho==T]cscopy_with_lesion_and_biopsy <- cscopy_with_lesion_and_biopsy[path_bun_t_metabolic==F & faci_bun_t_emergency==F]cscopy_with_lesion_and_biopsy %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 167 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_mod_sed 0.174
## 2 anes_bun_t_anes 0.0972
## 3 surg_bun_t_resect 0.0952
## 4 surg_bun_t_control 0.0851
## 5 surg_bun_t_hemorrhoid 0.0783
## 6 surg_bun_t_ligation 0.0783
## 7 path_bun_t_sex 0.0613
## 8 path_bun_t_testosterone 0.0613
## 9 surg_bun_t_breast 0.0608
## 10 medi_bun_t_stud 0.0602
## # ... with 157 more rowscscopy_with_lesion_and_biopsy %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.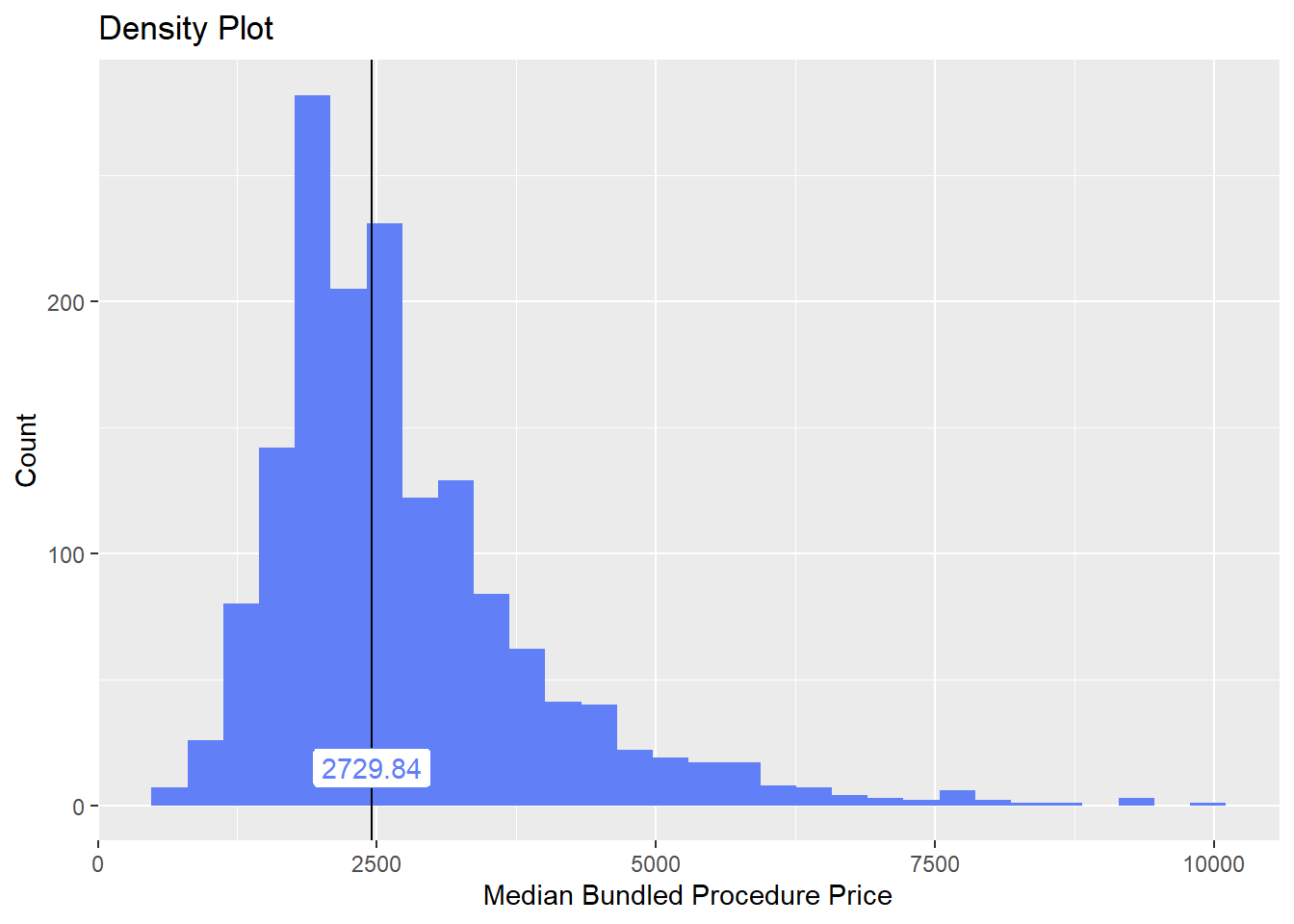
cscopy_with_lesion_and_biopsy %>% get_tag_density_information("anes_bun_t_mod_sed") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ anes_bun_t_mod_sed"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 263## $dist_plots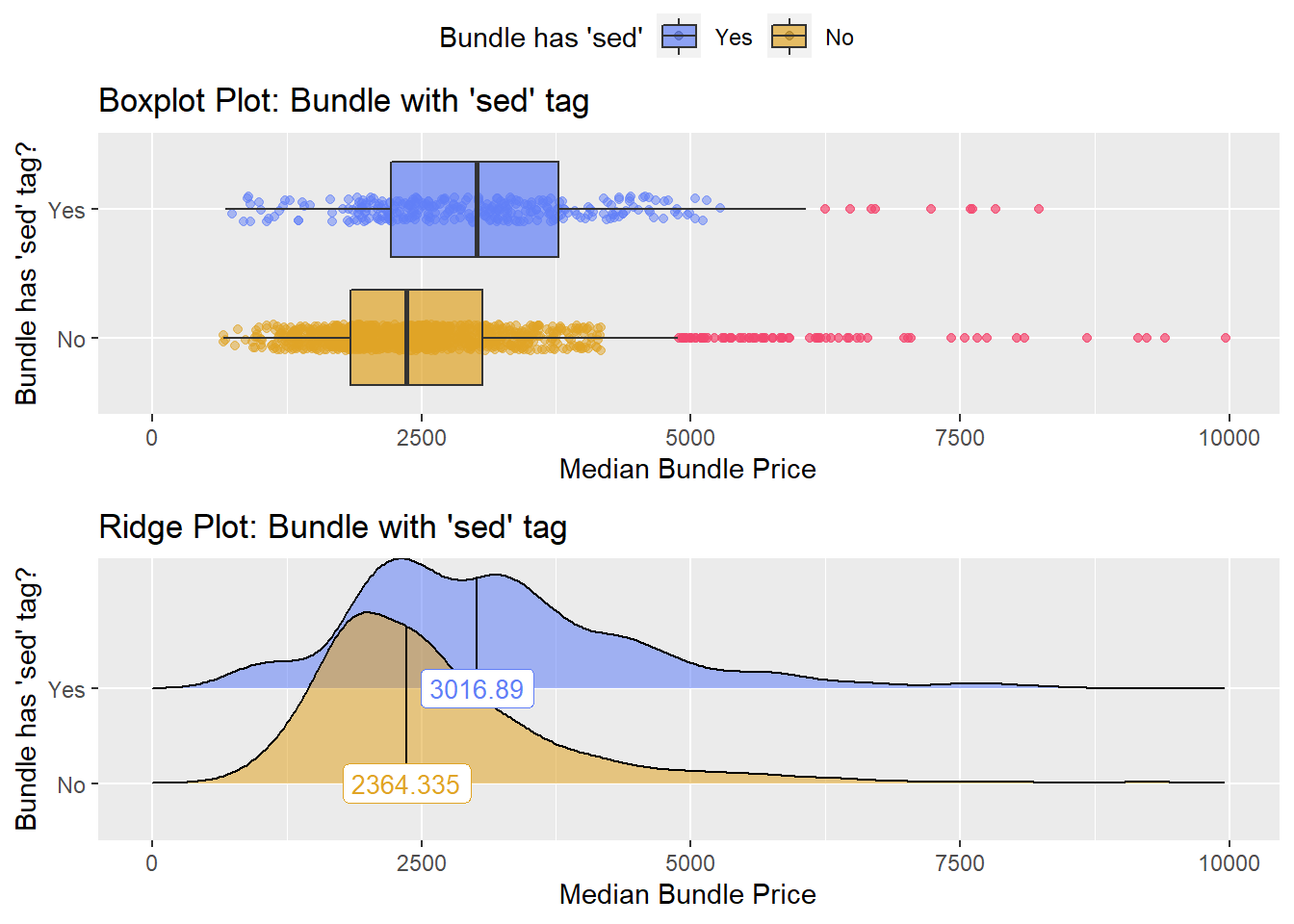
##
## $stat_tables there is strong evidence to split those groups into two.
there is strong evidence to split those groups into two.
cscopy_with_lesion_and_biopsy_and_mod_sed <- cscopy_with_lesion_and_biopsy[anes_bun_t_mod_sed==T]
cscopy_with_lesion_and_biopsy <- cscopy_with_lesion_and_biopsy[anes_bun_t_mod_sed==F]lets check the dist and correlated tags
cscopy_with_lesion_and_biopsy %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 146 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_resect 0.131
## 2 anes_bun_t_anes 0.111
## 3 path_bun_t_immunohisto 0.0857
## 4 path_bun_t_immuno 0.0837
## 5 path_bun_t_hormone 0.0738
## 6 path_bun_t_sex 0.0737
## 7 path_bun_t_testosterone 0.0737
## 8 surg_bun_t_breast 0.0731
## 9 faci_bun_t_est 0.0577
## 10 medi_bun_t_evaluat 0.0571
## # ... with 136 more rowscscopy_with_lesion_and_biopsy %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.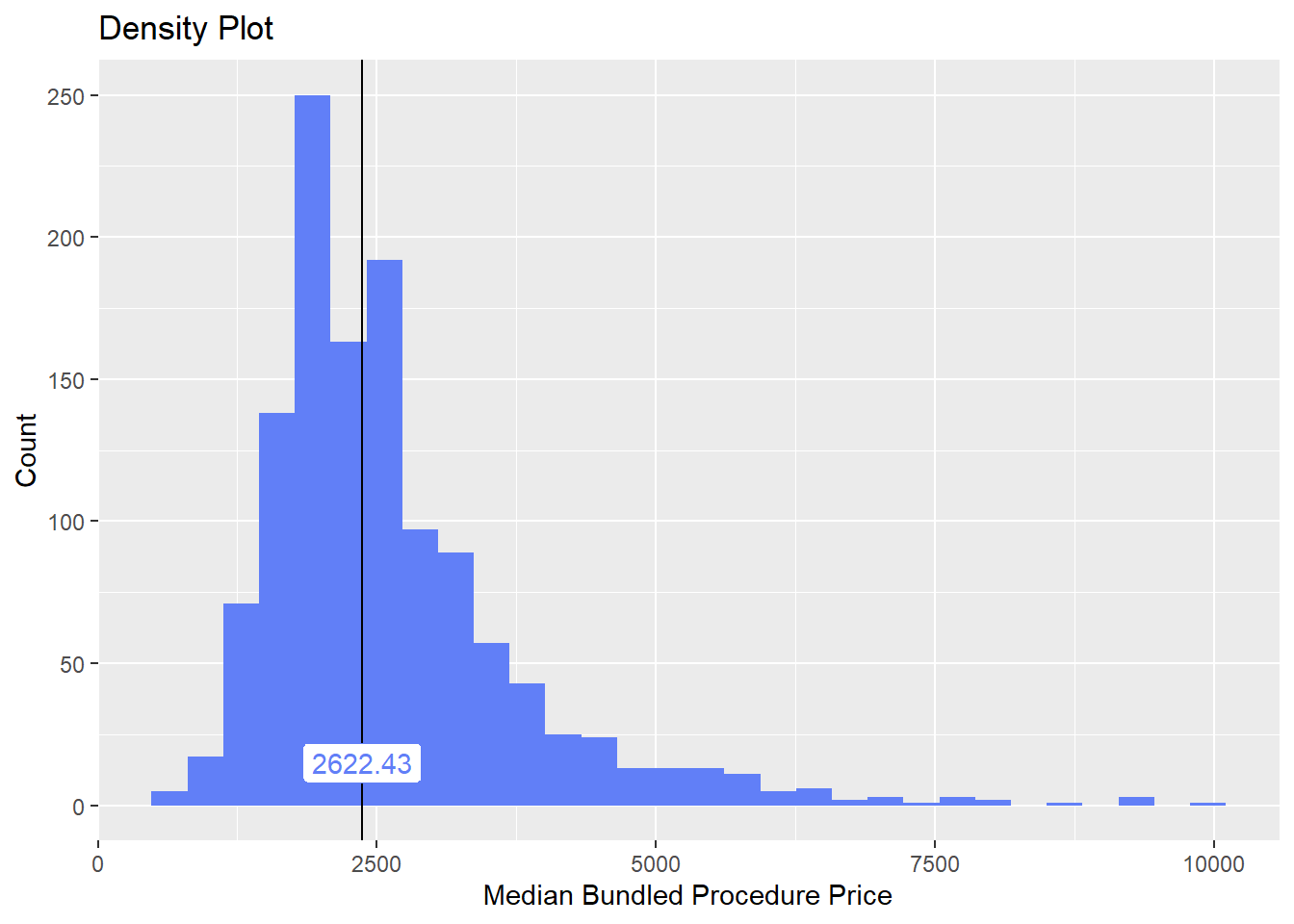
cscopy_with_lesion_and_biopsy <- cscopy_with_lesion_and_biopsy[surg_bun_t_resect==F & path_bun_t_immuno==F & path_bun_t_hormone==F & path_bun_t_sex==F & surg_bun_t_breast==F]cscopy_with_lesion_and_biopsy2 <- cscopy_with_lesion_and_biopsycscopy_with_lesion_and_biopsy_btbv4 <- cscopy_with_lesion_and_biopsy %>% btbv4()cscopy_with_lesion_and_biopsy_btbv4 <- cscopy_with_lesion_and_biopsy_btbv4 %>%
filter(tp_cnt_cnt>5) %>%
arrange(most_important_fac) %>%
select(doctor_str1,
doctor_str2,
doctor_npi_str1,
doctor_npi_str2,
most_important_fac,
most_important_fac_npi,
tp_med_med,
tp_med_surg,
tp_med_medi,
tp_med_path,
tp_med_radi,
tp_med_anes,
tp_med_faci,
tp_cnt_cnt) %>%
mutate(procedure_type = 1,
procedure_modifier = "Lesion Removal AND Biopsy")
cscopy_with_lesion_and_biopsy_btbv4## doctor_str1 doctor_str2 doctor_npi_str1 doctor_npi_str2
## 1: NOT SPECIFIED <NA> 0 <NA>
## 2: CHRISTOPHER CANALE <NA> 1598952145 <NA>
## 3: DAVID RAYBURN MOORE <NA> 1215969837 <NA>
## 4: BRETT W DOXEY <NA> 1386775682 <NA>
## 5: DAVID RAYBURN MOORE ERIC C JOHNSTON 1215969837 1407997273
## ---
## 191: PEDER J PEDERSEN <NA> 1568558211 <NA>
## 192: JASON C WILLS <NA> 1871689554 <NA>
## 193: HOLLY CLARK <NA> 1376639047 <NA>
## 194: TIMOTHY C HOLLINGSED <NA> 1790716181 <NA>
## 195: NOT SPECIFIED <NA> 0 <NA>
## most_important_fac most_important_fac_npi
## 1: BEAVER VALLEY HOSPITAL (BEAVER) 1174979108
## 2: BOUNTIFUL SURGERY CENTER (BOUNTIFUL) 1669415451
## 3: BOUNTIFUL SURGERY CENTER (BOUNTIFUL) 1669415451
## 4: BOUNTIFUL SURGERY CENTER (BOUNTIFUL) 1669415451
## 5: BOUNTIFUL SURGERY CENTER (BOUNTIFUL) 1669415451
## ---
## 191: WASATCH ENDOSCOPY CENTER LTD (SALT LAKE CITY) 1881701290
## 192: WASATCH ENDOSCOPY CENTER LTD (SALT LAKE CITY) 1881701290
## 193: WASATCH ENDOSCOPY CENTER LTD (SALT LAKE CITY) 1881701290
## 194: WASATCH FRONT SURGERY CENTER (WEST VALLEY CITY) 1063720365
## 195: WASATCH FRONT SURGERY CENTER (WEST VALLEY CITY) 1063720365
## tp_med_med tp_med_surg tp_med_medi tp_med_path tp_med_radi tp_med_anes
## 1: 3288.050 1661.53 0.00 544.00 0 550.960
## 2: 2943.600 2530.56 0.00 198.70 0 303.000
## 3: 2675.520 2530.56 0.00 273.66 0 260.000
## 4: 2597.905 2236.86 0.00 161.72 0 199.325
## 5: 2753.220 2563.51 0.00 136.00 0 0.000
## ---
## 191: 2519.240 1910.21 0.00 278.18 0 370.175
## 192: 2592.370 2091.71 0.00 221.84 0 260.000
## 193: 3027.790 2319.41 0.00 352.38 0 356.000
## 194: 3744.220 2658.94 88.20 997.08 0 0.000
## 195: 2296.840 1691.82 6.61 598.41 0 0.000
## tp_med_faci tp_cnt_cnt procedure_type procedure_modifier
## 1: 531.56 7 1 Lesion Removal AND Biopsy
## 2: 0.00 111 1 Lesion Removal AND Biopsy
## 3: 0.00 83 1 Lesion Removal AND Biopsy
## 4: 0.00 45 1 Lesion Removal AND Biopsy
## 5: 53.71 6 1 Lesion Removal AND Biopsy
## ---
## 191: 0.00 59 1 Lesion Removal AND Biopsy
## 192: 0.00 81 1 Lesion Removal AND Biopsy
## 193: 0.00 14 1 Lesion Removal AND Biopsy
## 194: 0.00 6 1 Lesion Removal AND Biopsy
## 195: 0.00 7 1 Lesion Removal AND Biopsy3. Even more break downs.
While these are valid break downs, they seems to complex for the average customer to shop for, so I am going to display them, but they will not be in the final tool.
cscopy_with_lesion_biopsy_injection %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 198 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_mod_sed 0.442
## 2 surg_bun_t_resect 0.264
## 3 path_bun_t_panel 0.152
## 4 path_bun_t_metabolic 0.148
## 5 path_bun_t_total 0.142
## 6 surg_bun_t_tags 0.141
## 7 medi_bun_t_emotional 0.141
## 8 medi_bun_t_stud 0.141
## 9 faci_bun_t_dept 0.140
## 10 faci_bun_t_emergency 0.140
## # ... with 188 more rowscscopy_with_lesion_biopsy_injection %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.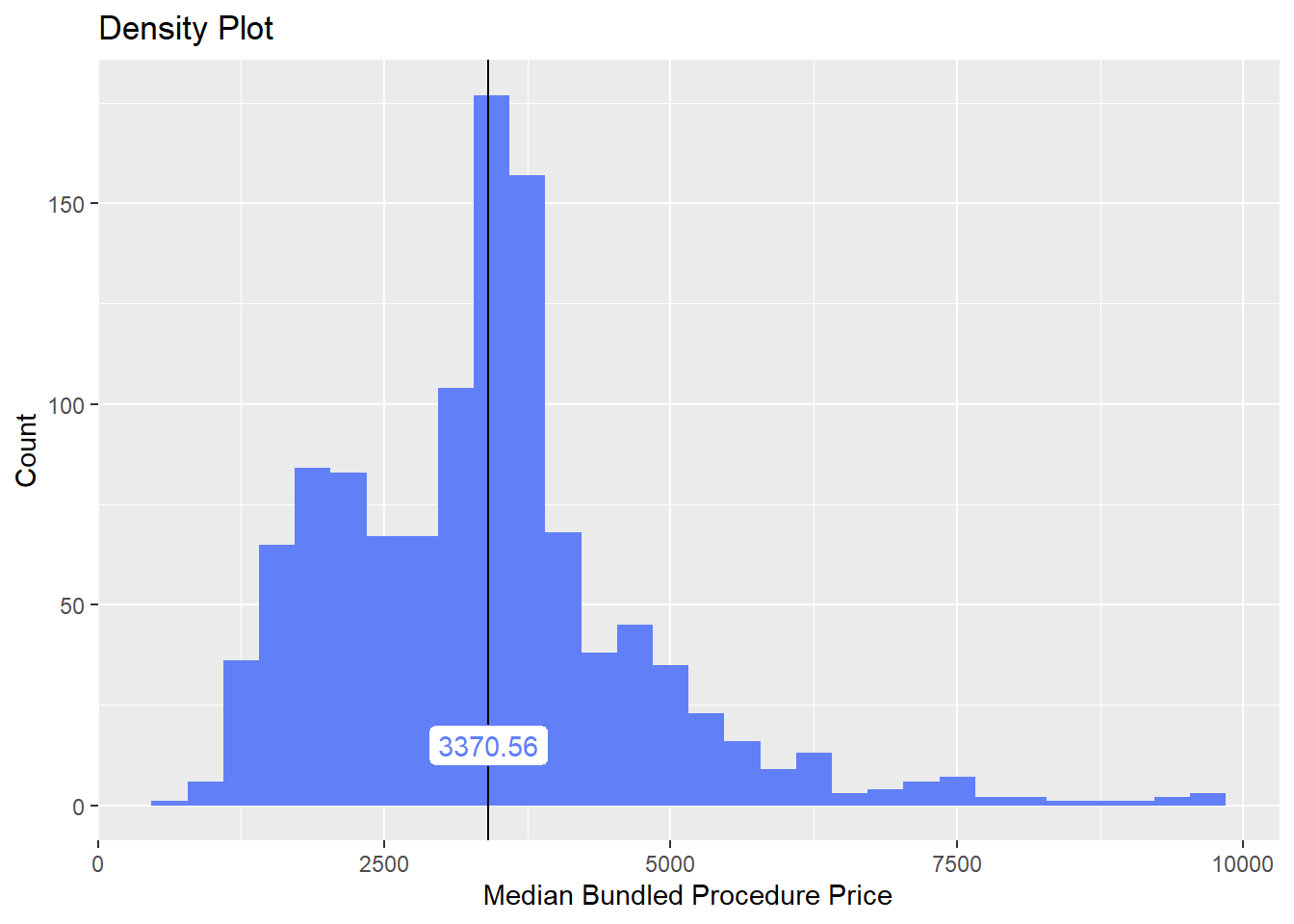
cscopy_with_lesion_biopsy_injection %>% get_tag_density_information("anes_bun_t_mod_sed") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ anes_bun_t_mod_sed"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 205## $dist_plots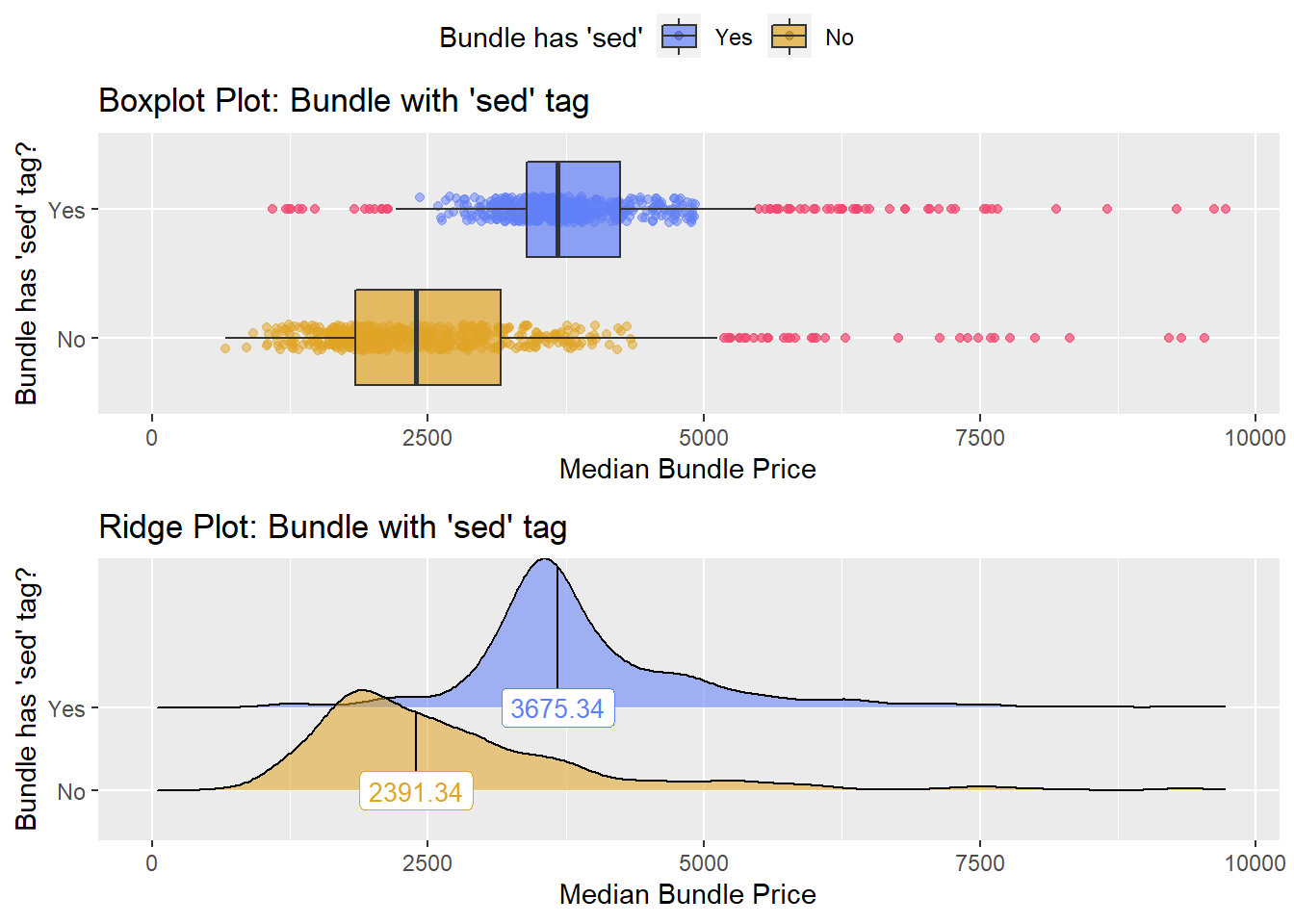
##
## $stat_tables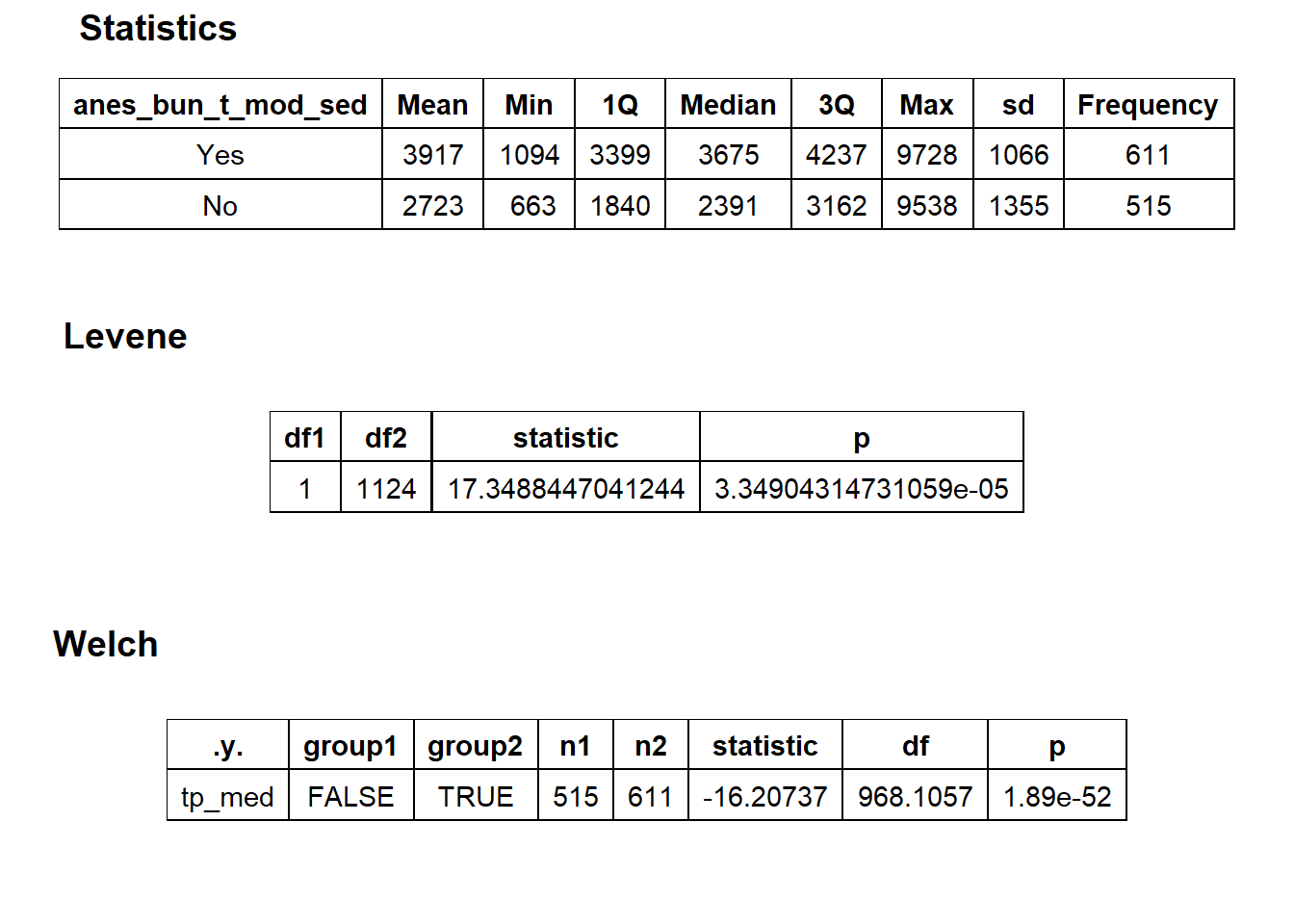
cscopy_with_lesion_biopsy_injection <- cscopy_with_lesion_biopsy_injection[surg_bun_t_resect==F] cscopy_with_lesion_biopsy_injection_mod_sed <- cscopy_with_lesion_biopsy_injection[anes_bun_t_mod_sed==T]
cscopy_with_lesion_biopsy_injection <- cscopy_with_lesion_biopsy_injection[anes_bun_t_mod_sed==F]cscopy_with_lesion_biopsy_injection %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 155 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_anes 0.305
## 2 path_bun_t_blood 0.300
## 3 path_bun_t_panel 0.264
## 4 path_bun_t_glucose 0.248
## 5 path_bun_t_metabolic 0.246
## 6 medi_bun_t_choline 0.244
## 7 medi_bun_t_device 0.243
## 8 medi_bun_t_draw 0.243
## 9 medi_bun_t_haloperidol 0.232
## 10 anes_bun_t_anesth 0.232
## # ... with 145 more rowscscopy_with_lesion_biopsy_injection %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.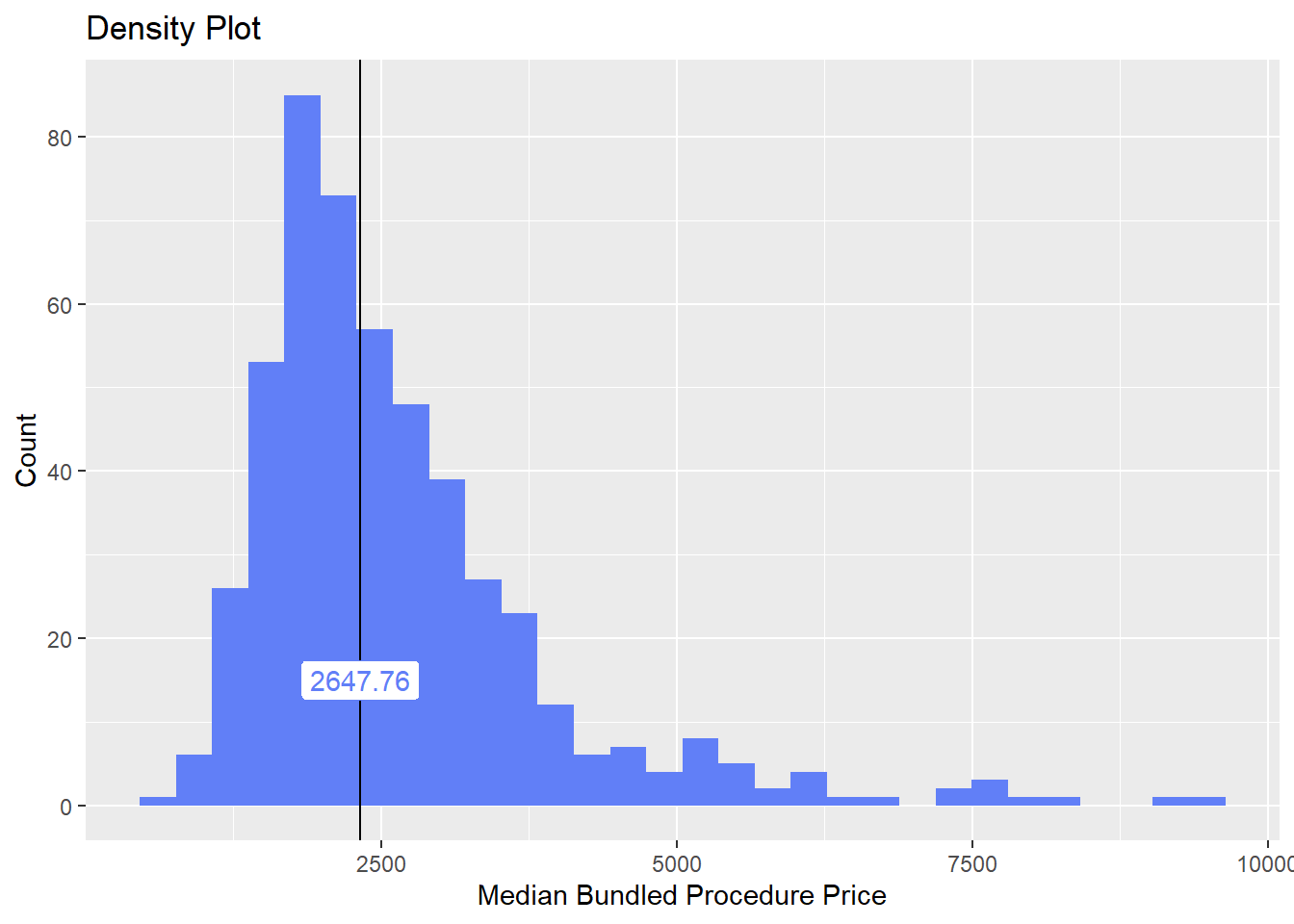
cscopy_with_lesion_biopsy_injection_mod_sed %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 121 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_tags 0.240
## 2 medi_bun_t_emotional 0.240
## 3 medi_bun_t_stud 0.240
## 4 faci_bun_t_dept 0.240
## 5 faci_bun_t_emergency 0.240
## 6 path_bun_t_psa 0.210
## 7 path_bun_t_total 0.192
## 8 path_bun_t_comprehen 0.181
## 9 path_bun_t_metabolic 0.181
## 10 path_bun_t_assay 0.162
## # ... with 111 more rowscscopy_with_lesion_biopsy_injection_mod_sed %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
D. Tissue pathology
I will now break down the Colonoscopy with a tissue pathology
lets look at the distribution and correlated tags
cscopy_with_tissue %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
cscopy_with_tissue %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 363 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_anesth 0.274
## 2 anes_bun_t_surg 0.261
## 3 medi_bun_t_sodium 0.238
## 4 anes_bun_t_anes 0.216
## 5 anes_bun_t_mod_sed 0.199
## 6 surg_bun_t_remov 0.197
## 7 surg_bun_t_remove 0.189
## 8 faci_bun_t_discharge 0.141
## 9 surg_bun_t_biopsy 0.139
## 10 surg_bun_t_appendectomy 0.138
## # ... with 353 more rowscscopy_with_tissue %>% get_tag_density_information("anes_bun_t_anesth") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ anes_bun_t_anesth"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 575## $dist_plots
##
## $stat_tables valid split, but not a large enough grouping to warrant own category.
valid split, but not a large enough grouping to warrant own category.
cscopy_with_tissue <- cscopy_with_tissue[anes_bun_t_anesth == F]lets look at the distribution and correlated tags
cscopy_with_tissue %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
cscopy_with_tissue %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 339 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_mod_sed 0.220
## 2 anes_bun_t_anes 0.175
## 3 faci_bun_t_discharge 0.149
## 4 faci_bun_t_initial 0.144
## 5 faci_bun_t_dept 0.141
## 6 faci_bun_t_emergency 0.141
## 7 medi_bun_t_normal 0.131
## 8 medi_bun_t_saline 0.131
## 9 medi_bun_t_solution 0.131
## 10 medi_bun_t_sodium 0.130
## # ... with 329 more rowscscopy_with_tissue %>% get_tag_density_information("anes_bun_t_mod_sed") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ anes_bun_t_mod_sed"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 81.4## $dist_plots
##
## $stat_tables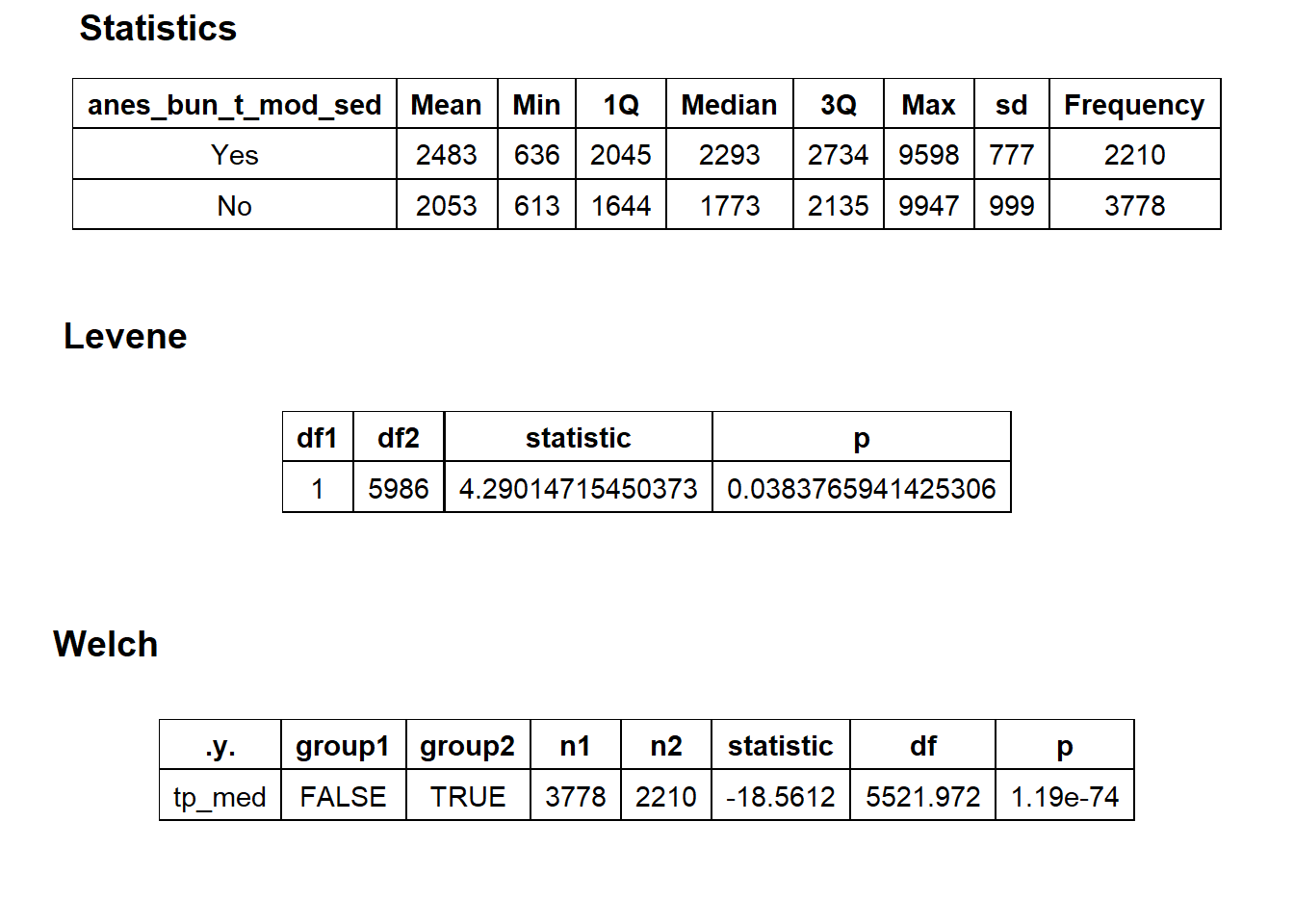
cscopy_with_tissue_with_mod_sed <- cscopy_with_tissue[anes_bun_t_mod_sed == T]
cscopy_with_tissue <- cscopy_with_tissue[anes_bun_t_mod_sed == F]cscopy_with_tissue %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
cscopy_with_tissue %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 295 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_anes 0.237
## 2 faci_bun_t_discharge 0.198
## 3 faci_bun_t_initial 0.191
## 4 medi_bun_t_normal 0.176
## 5 medi_bun_t_saline 0.176
## 6 medi_bun_t_solution 0.176
## 7 medi_bun_t_sodium 0.170
## 8 faci_bun_t_dept 0.169
## 9 faci_bun_t_emergency 0.169
## 10 faci_bun_t_care 0.162
## # ... with 285 more rowscscopy_with_tissue %>% get_tag_density_information("faci_bun_t_hospital") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ faci_bun_t_hospital"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 1140## $dist_plots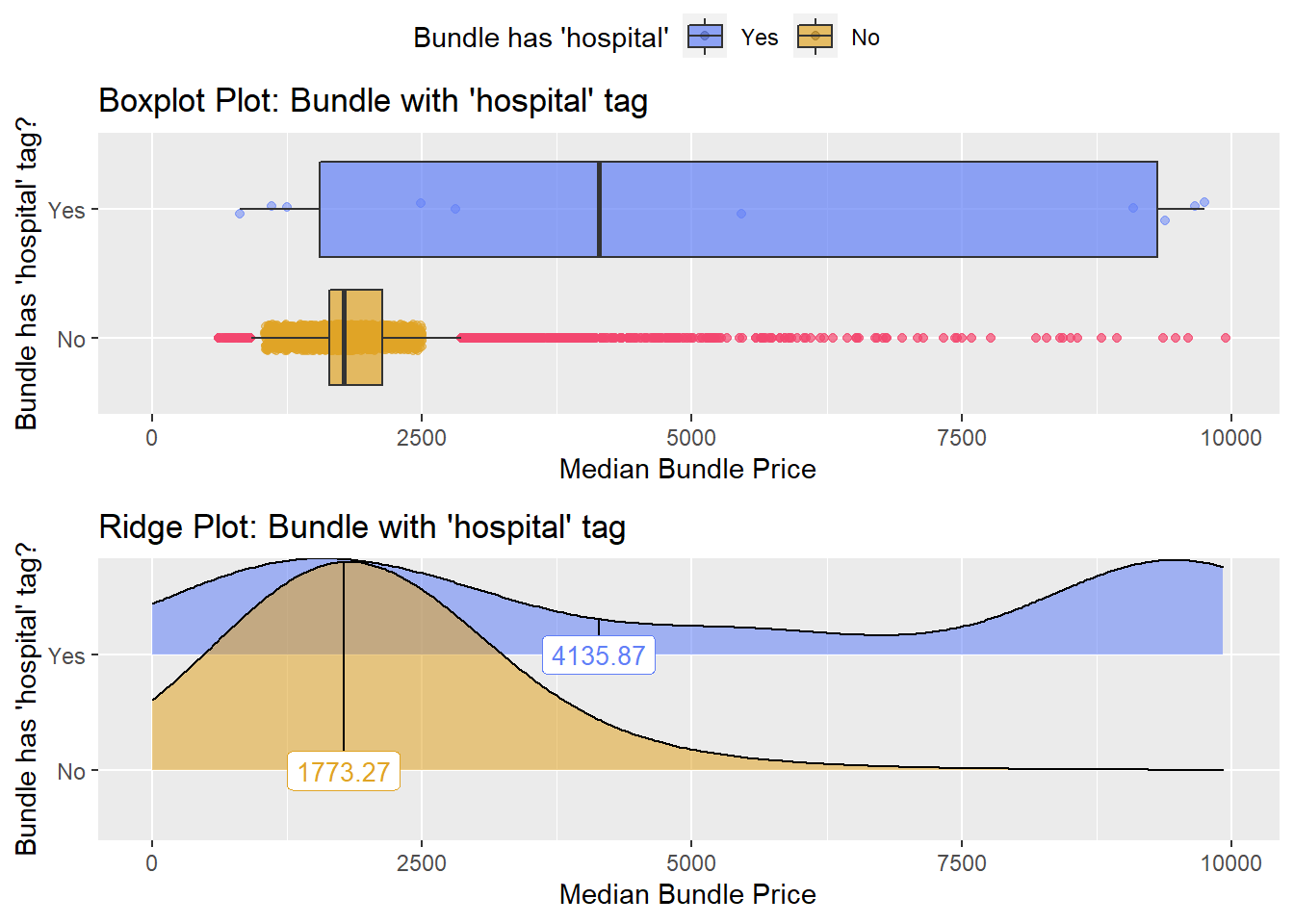
##
## $stat_tables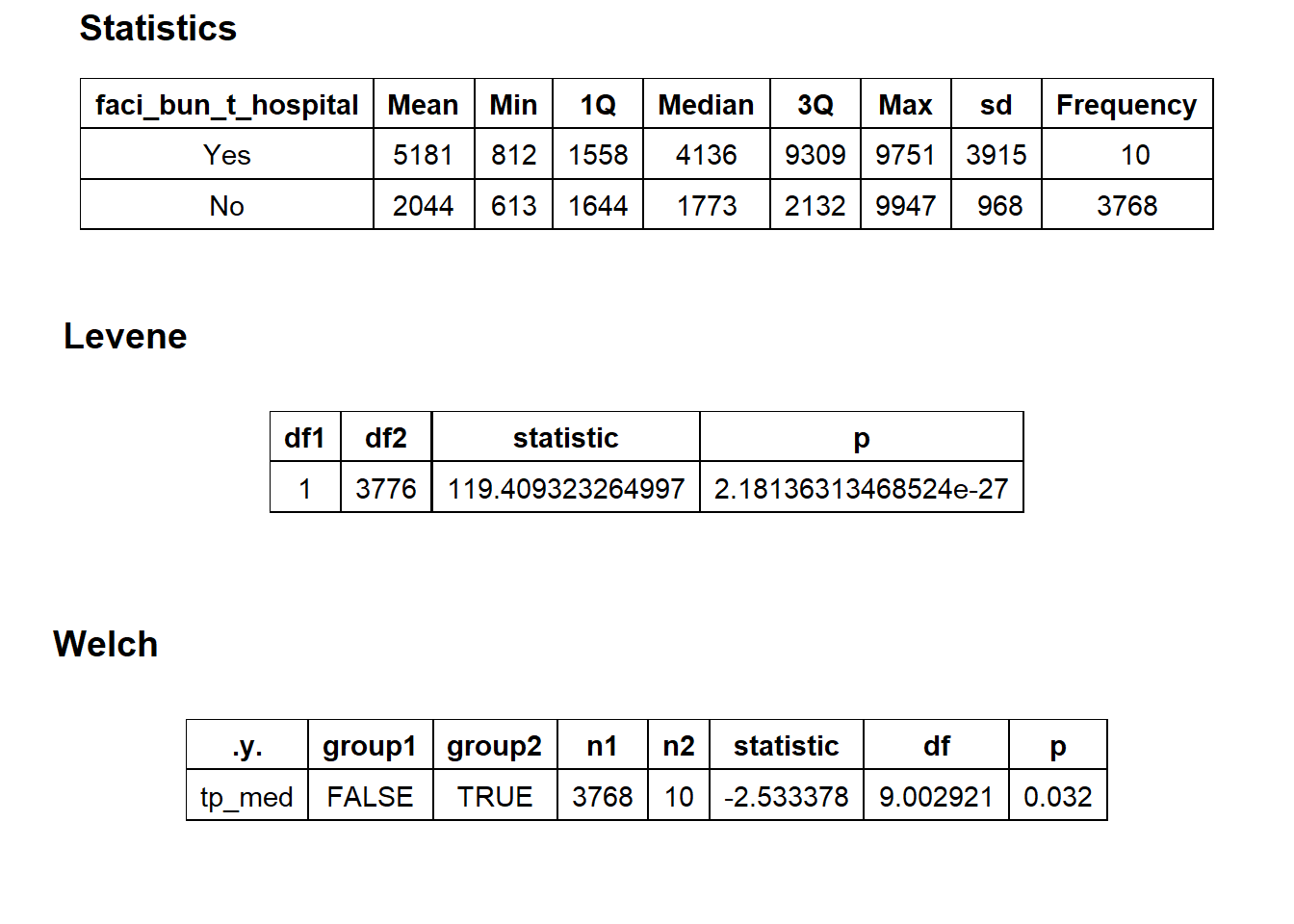
cscopy_with_tissue <- cscopy_with_tissue[faci_bun_t_initial==F & medi_bun_t_normal==F & faci_bun_t_discharge==F & faci_bun_t_emergency==F & faci_bun_t_hospital==F & medi_bun_t_sodium==F & surg_bun_t_resect==F & path_bun_t_panel==F & surg_bun_t_anal==F & surg_bun_t_destroy==F & medi_bun_t_wheezing==F & surg_bun_t_nasal == F & path_bun_t_hiv==F & path_bun_t_allerg==F & path_bun_t_pregnancy==F & surg_bun_t_dilat==F]cscopy_with_tissue %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.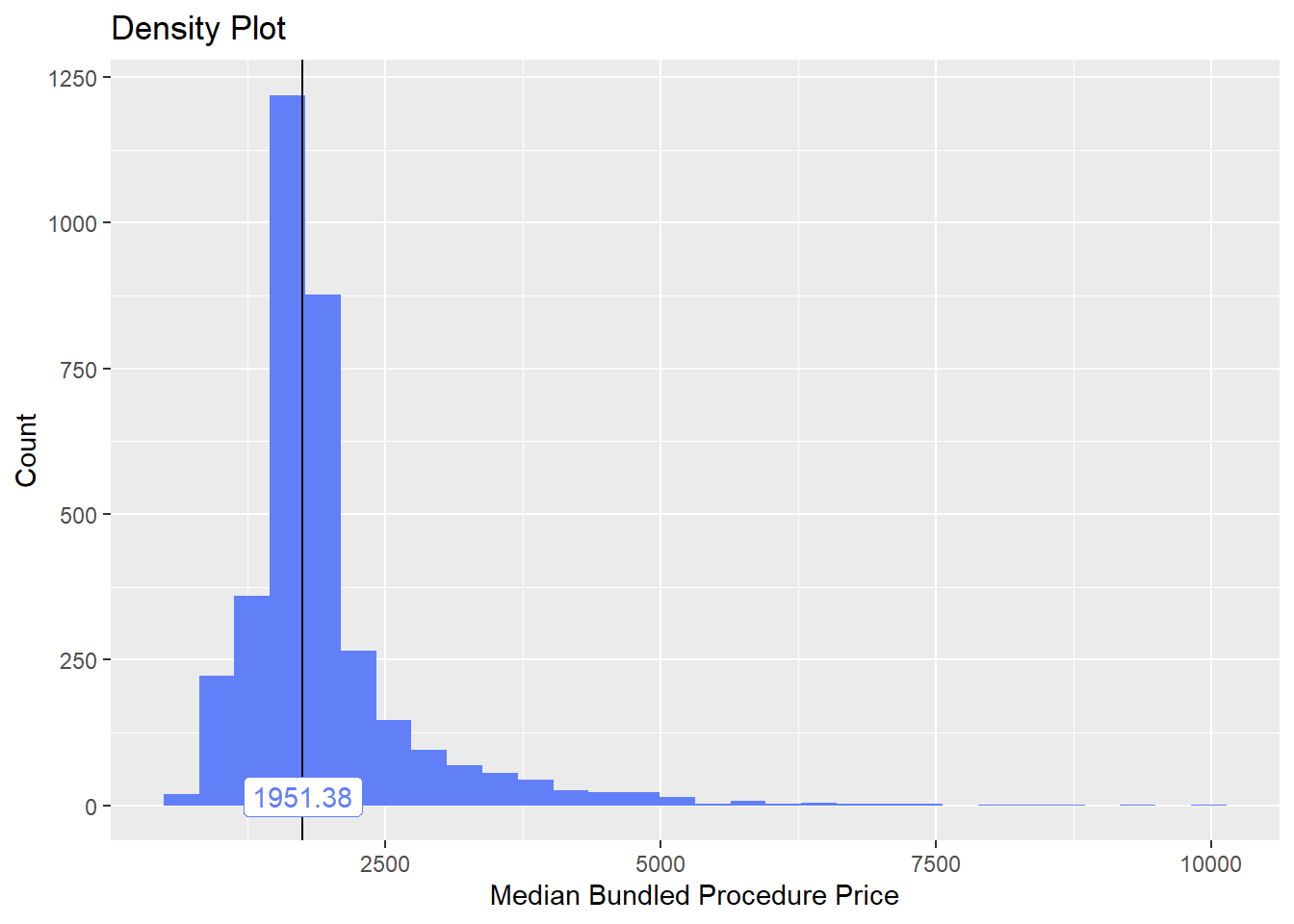
cscopy_with_tissue %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 211 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_anes 0.237
## 2 surg_bun_t_njx 0.0881
## 3 surg_bun_t_sub 0.0824
## 4 medi_bun_t_tracing 0.0642
## 5 faci_bun_t_est 0.0608
## 6 medi_bun_t_methylprednisolone 0.0604
## 7 medi_bun_t_prednisolone 0.0604
## 8 path_bun_t_gene 0.0604
## 9 surg_bun_t_control 0.0597
## 10 faci_bun_t_visit 0.0579
## # ... with 201 more rowscscopy_with_tissue_btbv4 <- cscopy_with_tissue %>% btbv4()cscopy_with_tissue_btbv4 <- cscopy_with_tissue_btbv4 %>%
filter(tp_cnt_cnt>5) %>%
arrange(most_important_fac) %>%
select(doctor_str1,
doctor_str2,
doctor_npi_str1,
doctor_npi_str2,
most_important_fac,
most_important_fac_npi,
tp_med_med,
tp_med_surg,
tp_med_medi,
tp_med_path,
tp_med_radi,
tp_med_anes,
tp_med_faci,
tp_cnt_cnt) %>%
mutate(procedure_type = 1,
procedure_modifier = "Tissue Pathology")
cscopy_with_tissue_btbv4## doctor_str1 doctor_str2 doctor_npi_str1
## 1: NOT SPECIFIED <NA> 0
## 2: MARIO DAMON LA GIGLIA <NA> 1013145697
## 3: CHRISTOPHER CANALE <NA> 1598952145
## 4: DAVID RAYBURN MOORE <NA> 1215969837
## 5: DAVID RAYBURN MOORE JOHN E ROBISON 1215969837
## ---
## 313: CHRISTOPHER IAN MAXWELL <NA> 1114107026
## 314: NOT SPECIFIED <NA> 0
## 315: DAVID BRADLEY TROWBRIDGE DOUGLAS MARR WOSETH 1134215809
## 316: TIMOTHY C HOLLINGSED <NA> 1790716181
## 317: DAVID OELSNER <NA> 1396760765
## doctor_npi_str2 most_important_fac
## 1: <NA> BEAVER VALLEY HOSPITAL (BEAVER)
## 2: <NA> BLUE MOUNTAIN HOSPITAL (BLANDING)
## 3: <NA> BOUNTIFUL SURGERY CENTER (BOUNTIFUL)
## 4: <NA> BOUNTIFUL SURGERY CENTER (BOUNTIFUL)
## 5: 1629187638 BOUNTIFUL SURGERY CENTER (BOUNTIFUL)
## ---
## 313: <NA> WASATCH ENDOSCOPY CENTER LTD (SALT LAKE CITY)
## 314: <NA> WASATCH ENDOSCOPY CENTER LTD (SALT LAKE CITY)
## 315: 1619978970 WASATCH ENDOSCOPY CENTER LTD (SALT LAKE CITY)
## 316: <NA> WASATCH FRONT SURGERY CENTER (WEST VALLEY CITY)
## 317: <NA> WASATCH FRONT SURGERY CENTER (WEST VALLEY CITY)
## most_important_fac_npi tp_med_med tp_med_surg tp_med_medi tp_med_path
## 1: 1174979108 3070.698 1580.175 14.5200 384.960
## 2: 1558513812 2086.440 1357.330 0.0000 212.120
## 3: 1669415451 1774.150 1614.010 0.0000 93.390
## 4: 1669415451 2034.560 1586.510 0.0000 99.455
## 5: 1669415451 2555.180 1675.820 0.0000 332.760
## ---
## 313: 1881701290 1835.383 1500.368 35.0125 114.915
## 314: 1881701290 1742.380 1402.350 0.0000 80.030
## 315: 1881701290 2428.740 1740.990 0.0000 261.820
## 316: 1063720365 2804.175 2244.540 88.2000 471.435
## 317: 1063720365 2005.770 1280.000 0.0000 725.770
## tp_med_radi tp_med_anes tp_med_faci tp_cnt_cnt procedure_type
## 1: 0 498.3950 515.02 52 1
## 2: 0 0.0000 516.99 7 1
## 3: 0 276.7500 0.00 71 1
## 4: 0 353.5000 112.01 132 1
## 5: 0 400.0000 146.60 6 1
## ---
## 313: 0 185.0875 0.00 42 1
## 314: 0 260.0000 0.00 16 1
## 315: 0 320.0000 105.93 6 1
## 316: 0 0.0000 0.00 13 1
## 317: 0 0.0000 0.00 6 1
## procedure_modifier
## 1: Tissue Pathology
## 2: Tissue Pathology
## 3: Tissue Pathology
## 4: Tissue Pathology
## 5: Tissue Pathology
## ---
## 313: Tissue Pathology
## 314: Tissue Pathology
## 315: Tissue Pathology
## 316: Tissue Pathology
## 317: Tissue Pathologycscopy_final <- cscopy_btbv4 %>%
rbind(cscopy_with_lesion_btbv4) %>%
rbind(cscopy_with_lesion_and_biopsy_btbv4) %>%
rbind(cscopy_with_tissue_btbv4)cscopy_final %>% saveRDS("cscopy_final.RDS")cscopy_final %>% nrow()## [1] 1012cscopy_final %>% count(doctor_str2) %>% arrange(desc(n))## doctor_str2 n
## 1: <NA> 853
## 2: MOHAMMAD M ALSOLAIMAN 5
## 3: PETER C. O. FENTON 5
## 4: MATTHEW EDWARD FEURER 4
## 5: JOHN E ROBISON 3
## ---
## 114: VERNON J COOLEY 1
## 115: WALSTIR H FONSECA 1
## 116: WALTER H. REICHERT 1
## 117: WAYNE W MORTENSEN 1
## 118: ZAHABIA TAHER GANDHI 1946 bundles. 71 rows
cscopy_final_bq <- cscopy_final[,
primary_doctor := pmap(.l=list(doctor_npi1=doctor_npi_str1,
doctor_npi2=doctor_npi_str2,
class_reqs="Internal Medicine|||Colon & Rectal Surgery",
specialization_reqs="Gastroenterology"),
.f=calculate_primary_doctor) %>% as.character()
] %>%
#Filter out any procedures where our doctors fail both criteria.
.[!(primary_doctor %in% c("BOTH_DOC_FAIL_CRIT", "TWO_FIT_ALL_SPECS", "NONE_FIT_SPEC_REQ"))] %>%
.[,primary_doctor_npi := fifelse(primary_doctor==doctor_str1,
doctor_npi_str1,
doctor_npi_str2)]## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "CASEY RAY OWENS"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "MOHAMMAD M ALSOLAIMAN" "DAVID J FRANTZ"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "CASEY RAY OWENS"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "CASEY RAY OWENS"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## character(0)## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "BRYCE DONALD HASLEM" "BRETT L THORPE"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "THOMAS A DICKINSON" "CHAD BARRETT KAWA"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "MARTIN I RADWIN"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## character(0)## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "DOUGLAS GRAHAM ADLER"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## character(0)## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "CASEY RAY OWENS"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "FAIZ AHMED SHAKIR"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "ALLEN CLARK GUNNERSON"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "ALLEN CLARK GUNNERSON"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "KURT O BODILY" "BRETT L THORPE"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "ALAN B ERDMANN"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "BRETT L THORPE" "CHAD BARRETT KAWA"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "VIKRAM GARG"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "KURT O BODILY"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "DAVID J FRANTZ"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "PETER C. O. FENTON"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "CHRISTOPHER CANALE"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "ROGER L SIDDOWAY"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "CHAD BARRETT KAWA"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "MATTHEW EDWARD FEURER"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "ROBERT G. JONES"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "EDWARD J FRECH"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "EDWARD J FRECH"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "PETER C. O. FENTON"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "JEFFREY S POOLE"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "DAN A COLLINS"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "MELVIN DEN KUWAHARA"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "DARCIE REASONER GORMAN"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "VIKRAM GARG"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "MATTHEW EDWARD FEURER"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "CHAD BARRETT KAWA"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "BRYCE DONALD HASLEM" "BRETT L THORPE"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used## [1] "multiple meet class req"
## [1] "BRIEN REX MILLER"## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be used
## Warning in if (!is.na(class_reqs)) {: the condition has length > 1 and only the
## first element will be usedcscopy_final_bq <- cscopy_final_bq[,.(
primary_doctor,
primary_doctor_npi,
most_important_fac ,
most_important_fac_npi,
procedure_type,
procedure_modifier,
tp_med_med,
tp_med_surg,
tp_med_medi,
tp_med_path,
tp_med_radi,
tp_med_anes,
tp_med_faci,
ingest_date = Sys.Date()
)]bq_table_upload(x=procedure_table, values= cscopy_final_bq, create_disposition='CREATE_IF_NEEDED', write_disposition='WRITE_APPEND')