Birth
Introduction
In this section we will: * identify which Delivery procedures were excluded and why * identify which different Delivery subgroups were created and why
If you have questions or concerns about this data please contact Alexander Nielson (alexnielson@utah.gov)
Load Libraries
Load Libraries
library(data.table)
library(tidyverse)## Warning: replacing previous import 'vctrs::data_frame' by 'tibble::data_frame'
## when loading 'dplyr'library(stringi)
library(ggridges)
library(broom)
library(disk.frame)
library(RecordLinkage)
library(googlesheets4)
library(bigrquery)
library(DBI)
devtools::install_github("utah-osa/hcctools2", upgrade="never" )
library(hcctools2)Establish color palettes
cust_color_pal1 <- c(
"Anesthesia" = "#f3476f",
"Facility" = "#e86a33",
"Medicine Proc" = "#e0a426",
"Pathology" = "#77bf45",
"Radiology" = "#617ff7",
"Surgery" = "#a974ee"
)
cust_color_pal2 <- c(
"TRUE" = "#617ff7",
"FALSE" = "#e0a426"
)
cust_color_pal3 <- c(
"above avg" = "#f3476f",
"avg" = "#77bf45",
"below avg" = "#a974ee"
)
fac_ref_regex <- "(UTAH)|(IHC)|(HOSP)|(HOSPITAL)|(CLINIC)|(ANESTH)|(SCOPY)|(SURG)|(LLC)|(ASSOC)|(MEDIC)|(CENTER)|(ASSOCIATES)|(U OF U)|(HEALTH)|(OLOGY)|(OSCOPY)|(FAMILY)|(VAMC)|(SLC)|(SALT LAKE)|(CITY)|(PROVO)|(OGDEN)|(ENDO )|( VALLEY)|( REGIONAL)|( CTR)|(GRANITE)|( INSTITUTE)|(INSTACARE)|(WASATCH)|(COUNTY)|(PEDIATRIC)|(CORP)|(CENTRAL)|(URGENT)|(CARE)|(UNIV)|(ODYSSEY)|(MOUNTAINSTAR)|( ORTHOPEDIC)|(INSTITUT)|(PARTNERSHIP)|(PHYSICIAN)|(CASTLEVIEW)|(CONSULTING)|(MAGEMENT)|(PRACTICE)|(EMERGENCY)|(SPECIALISTS)|(DIVISION)|(GUT WHISPERER)|(INTERMOUNTAIN)|(OBGYN)"Connect to GCP database
bigrquery::bq_auth(path = 'D:/gcp_keys/healthcare-compare-prod-95b3b7349c32.json')
# set my project ID and dataset name
project_id <- 'healthcare-compare-prod'
dataset_name <- 'healthcare_compare'
con <- dbConnect(
bigrquery::bigquery(),
project = project_id,
dataset = dataset_name,
billing = project_id
)Get NPI table
query <- paste0("SELECT npi, clean_name, osa_group, osa_class, osa_specialization
FROM `healthcare-compare-prod.healthcare_compare.npi_full`")
#bq_project_query(billing, query) # uncomment to determine billing price for above query.
npi_full <- dbGetQuery(con, query) %>%
data.table()get a subset of the NPI providers based upon taxonomy groups
gs4_auth(email="alexnielson@utah.gov")surgery <- read_sheet("https://docs.google.com/spreadsheets/d/1GY8lKwUJuPHtpUl4EOw9eriLUDG9KkNWrMbaSnA5hOU/edit#gid=0",
sheet="major_surgery") %>% as.data.table## Auto-refreshing stale OAuth token.## Reading from "Doctor Types to Keep"## Range "'major_surgery'"surgery <- surgery[is.na(Remove) ] %>% .[["NUCC Classification"]] npi_prov_pair <- npi_full[osa_class %in% surgery] %>%
.[,.(npi=npi,
clean_name = clean_name
)
]Load Data
bun_proc <- disk.frame("full_apcd.df")Birth
59610, 59612, 59614 - vbac delivery 59510, 59514, 59515 - cesarean delivery 59400, 59409, 59410 - Obstetrical care
omit:
d & c after delivery
birth_vag <- bun_proc[surg_bun_t_obstetrical ==T & surg_bun_t_care==T]
birth_csec <- bun_proc[surg_bun_t_delivery==T & surg_bun_t_cesarean==T]
birth_vbac <- bun_proc[surg_bun_t_delivery==T & stri_detect_regex(surg_bun_descr,"vbac")==T]vaginal birth
birth_vag <- birth_vag[,`:=`(
surg_sp_name_clean = surg_sp_npi %>% map_chr(get_npi_standard_name),
surg_bp_name_clean = surg_bp_npi %>% map_chr(get_npi_standard_name),
medi_sp_name_clean = medi_sp_npi %>% map_chr(get_npi_standard_name),
medi_bp_name_clean = medi_bp_npi %>% map_chr(get_npi_standard_name),
radi_sp_name_clean = radi_sp_npi %>% map_chr(get_npi_standard_name),
radi_bp_name_clean = radi_bp_npi %>% map_chr(get_npi_standard_name),
path_sp_name_clean = path_sp_npi %>% map_chr(get_npi_standard_name),
path_bp_name_clean = path_bp_npi %>% map_chr(get_npi_standard_name),
anes_sp_name_clean = anes_sp_npi %>% map_chr(get_npi_standard_name),
anes_bp_name_clean = anes_bp_npi %>% map_chr(get_npi_standard_name),
faci_sp_name_clean = faci_sp_npi %>% map_chr(get_npi_standard_name),
faci_bp_name_clean = faci_bp_npi %>% map_chr(get_npi_standard_name)
)]birth_vag %>% saveRDS("birth_vag.RDS")birth_vag <- birth_vag[cnt>5 & faci_bun_sum_med > 1000]We will now examine the vaginal birth deliveries’s median price distribution and cost correlated tags.
birth_vag[cnt>5 & faci_bun_sum_med > 1000]%>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
birth_vag[cnt>5 & faci_bun_sum_med > 1000]%>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 307 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.318
## 2 surg_bun_t_abd 0.268
## 3 surg_bun_t_abdom 0.268
## 4 surg_bun_t_abdomen 0.268
## 5 surg_bun_t_explora 0.268
## 6 anes_bun_t_abdom 0.246
## 7 faci_bun_t_care 0.242
## 8 faci_bun_t_initial 0.239
## 9 anes_bun_t_lower 0.237
## 10 faci_bun_t_hospital 0.236
## # ... with 297 more rowsbirth_vag %>% get_tag_density_information("surg_bun_t_abdomen") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_abdomen"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 4280## $dist_plots
##
## $stat_tables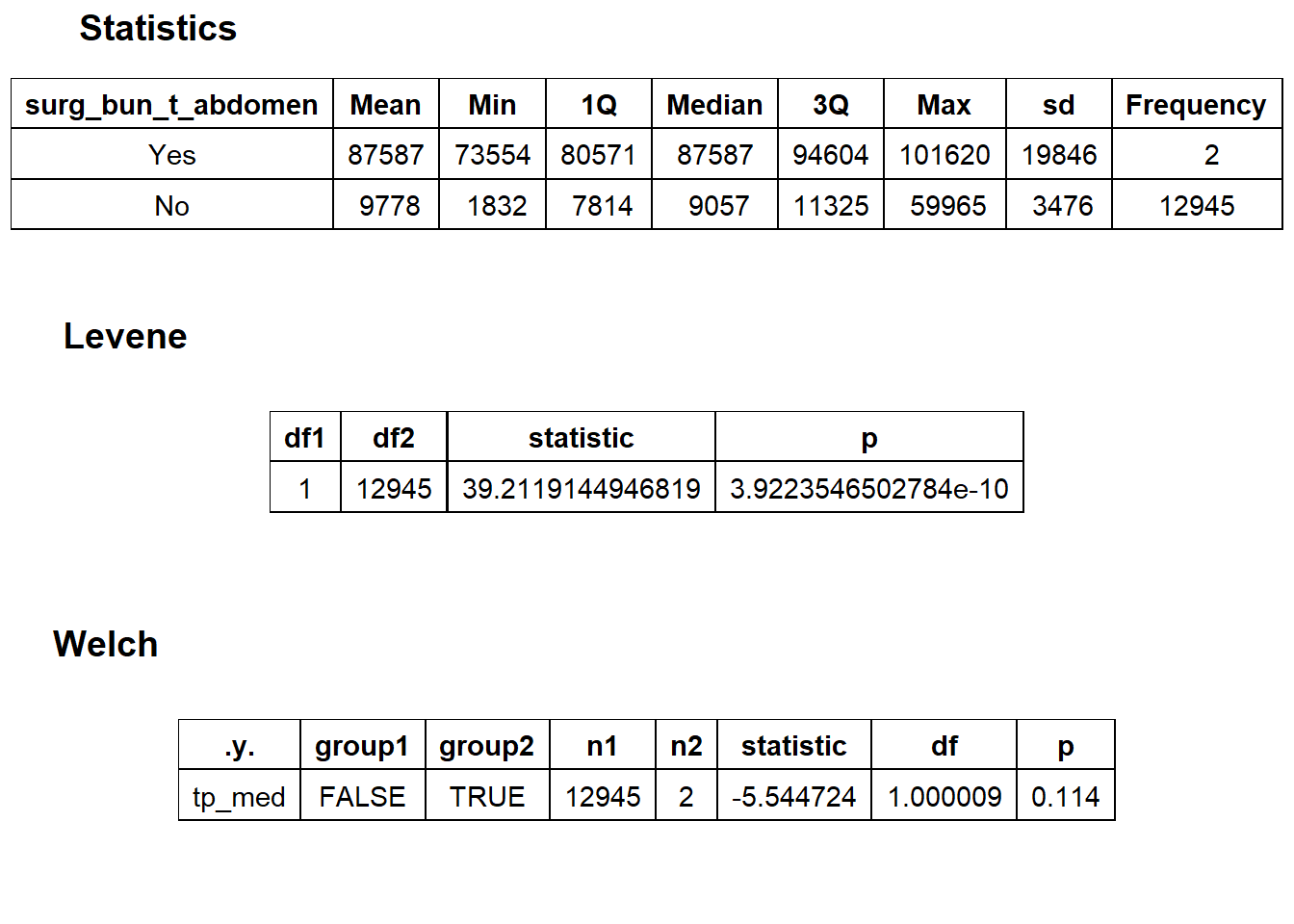
There are only two with a exploratory abdomen surgery code, so we will omit them from the standard.
birth_vag <- birth_vag[surg_bun_t_abdomen==F]We will now examine the vaginal birth deliveries’s median price distribution and cost correlated tags.
birth_vag%>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.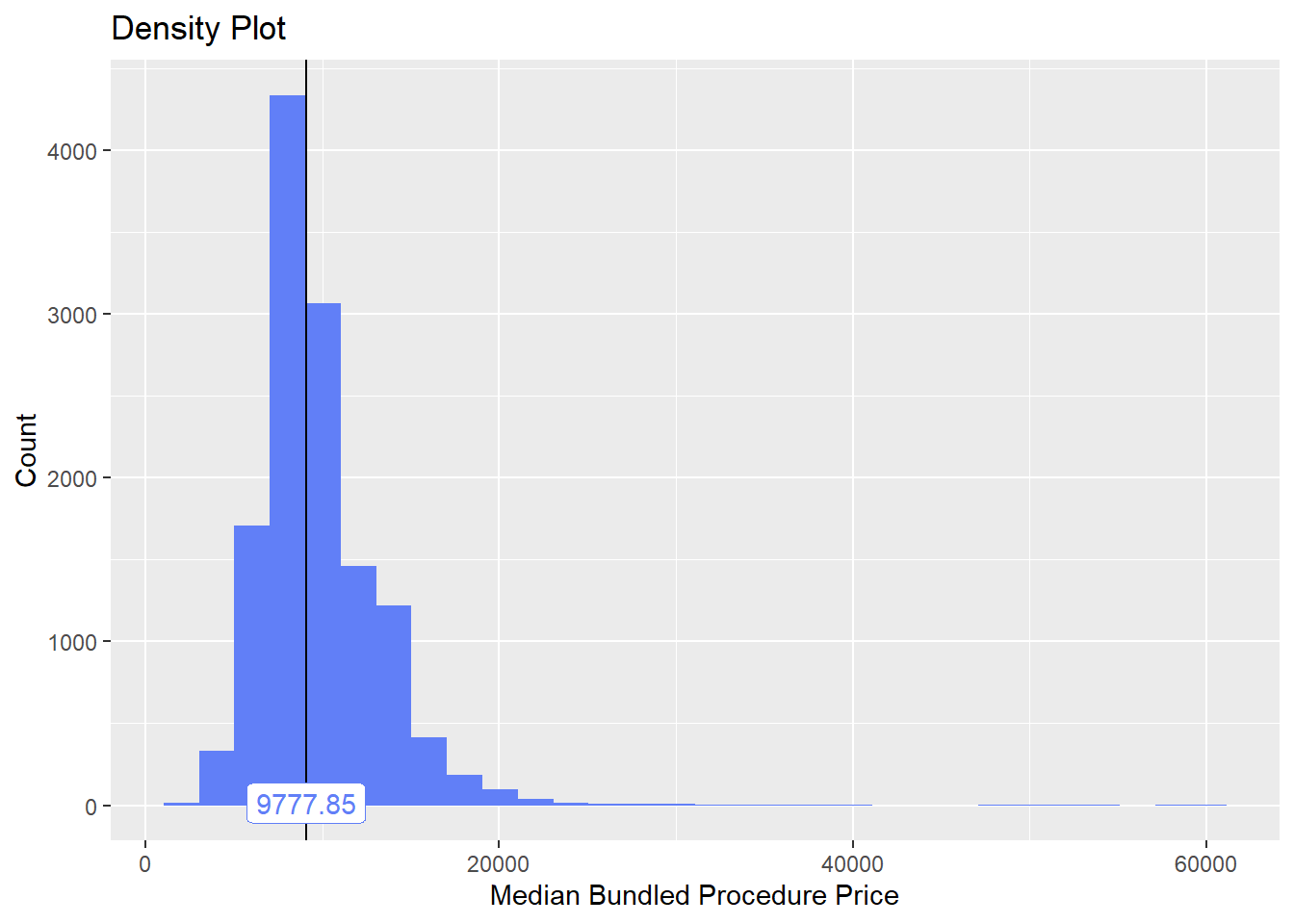
birth_vag%>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 300 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.304
## 2 faci_bun_t_care 0.234
## 3 faci_bun_t_hospital 0.229
## 4 faci_bun_t_initial 0.225
## 5 faci_bun_t_dept 0.218
## 6 faci_bun_t_emergency 0.218
## 7 faci_bun_t_visit 0.209
## 8 faci_bun_t_subsequent 0.195
## 9 anes_bun_t_anes 0.180
## 10 anes_bun_t_anesth 0.180
## # ... with 290 more rowsIt now looks like the main differece has to do with the facility type. ie: Initial hospital care Subsequent observation care Subsequent hospital care Emergency dept visit
birth_vag %>% get_tag_density_information("faci_bun_t_initial") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ faci_bun_t_initial"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 738## $dist_plots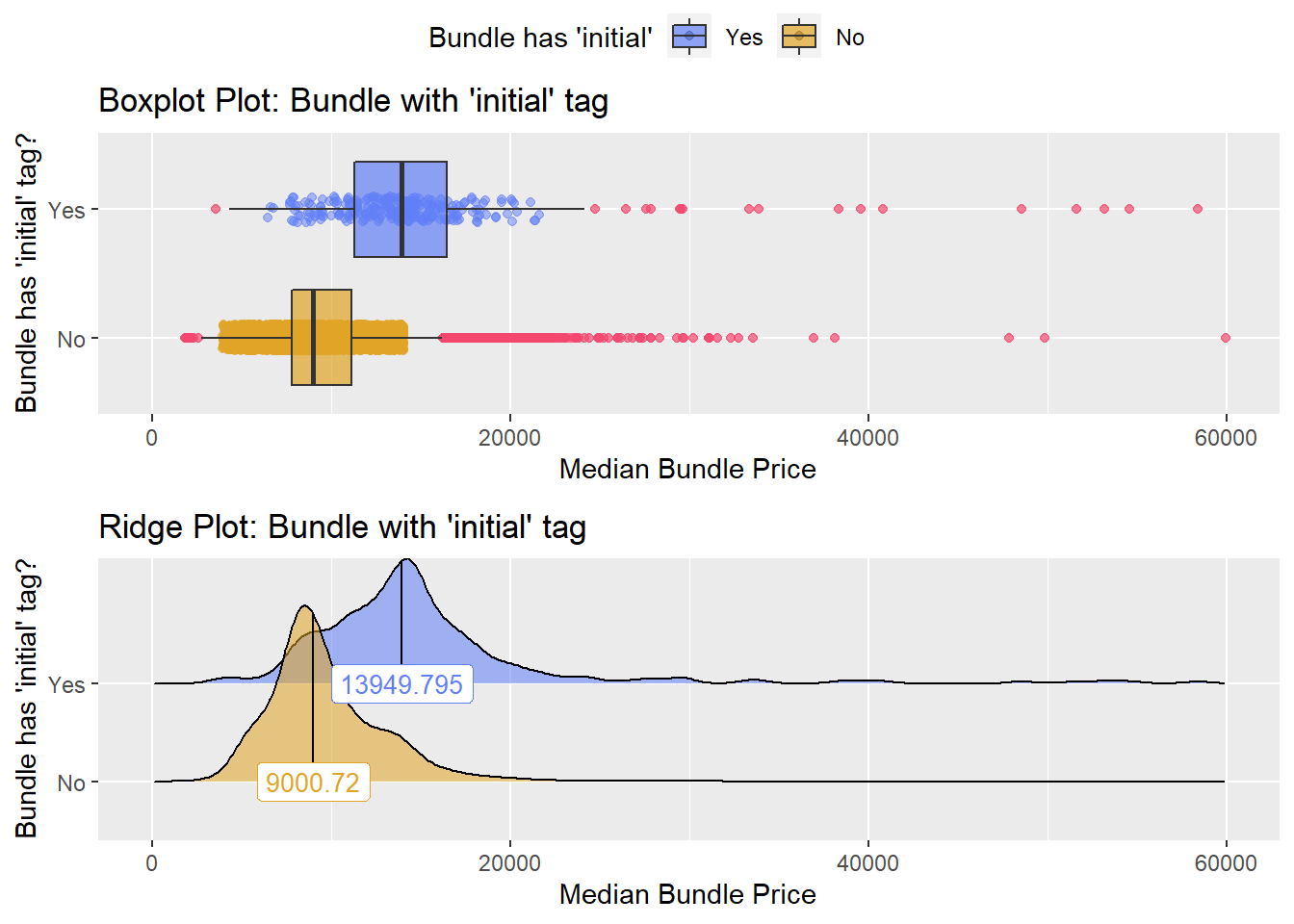
##
## $stat_tables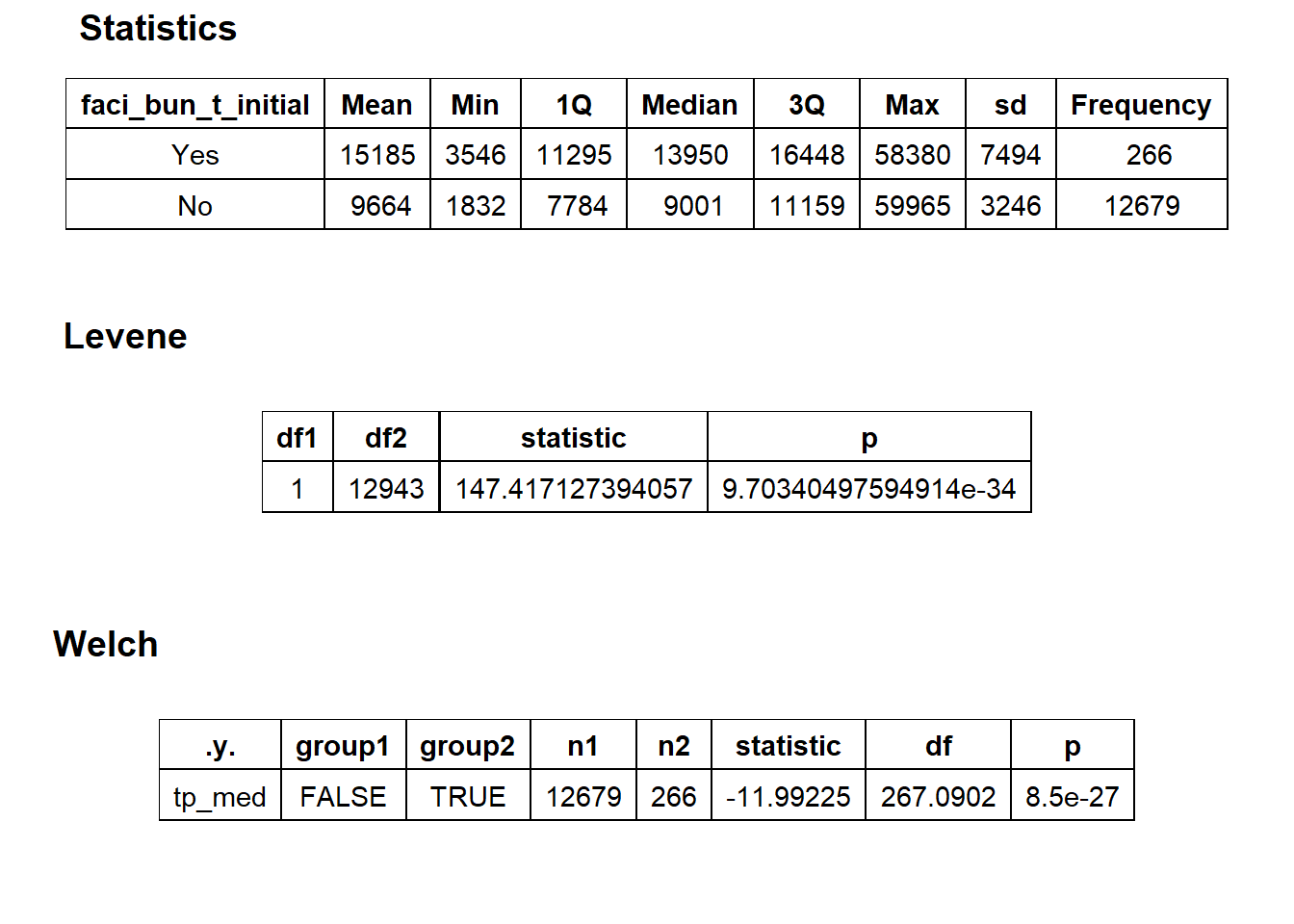
birth_vag %>% get_tag_density_information("faci_bun_t_emergency") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ faci_bun_t_emergency"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 467## $dist_plots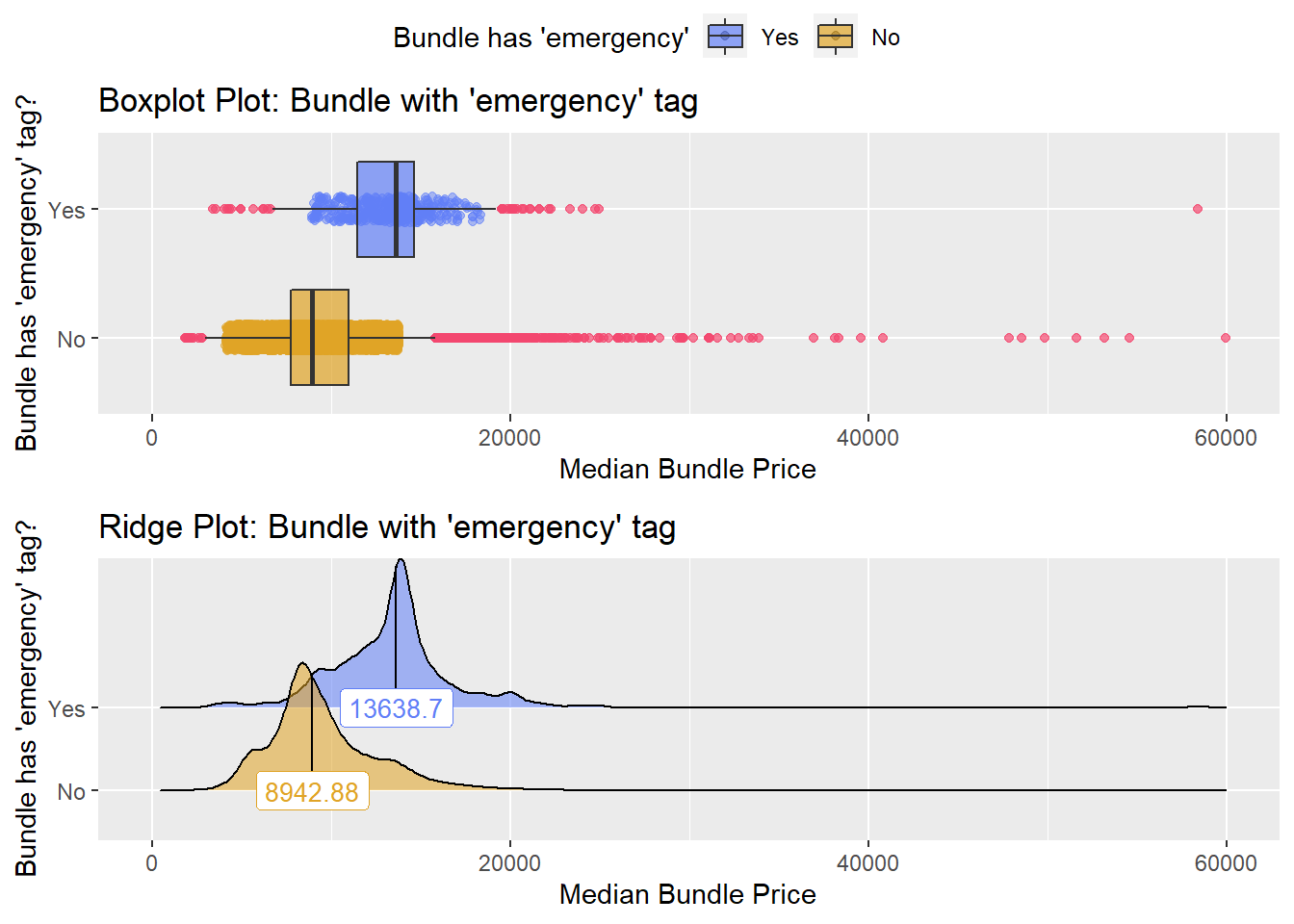
##
## $stat_tables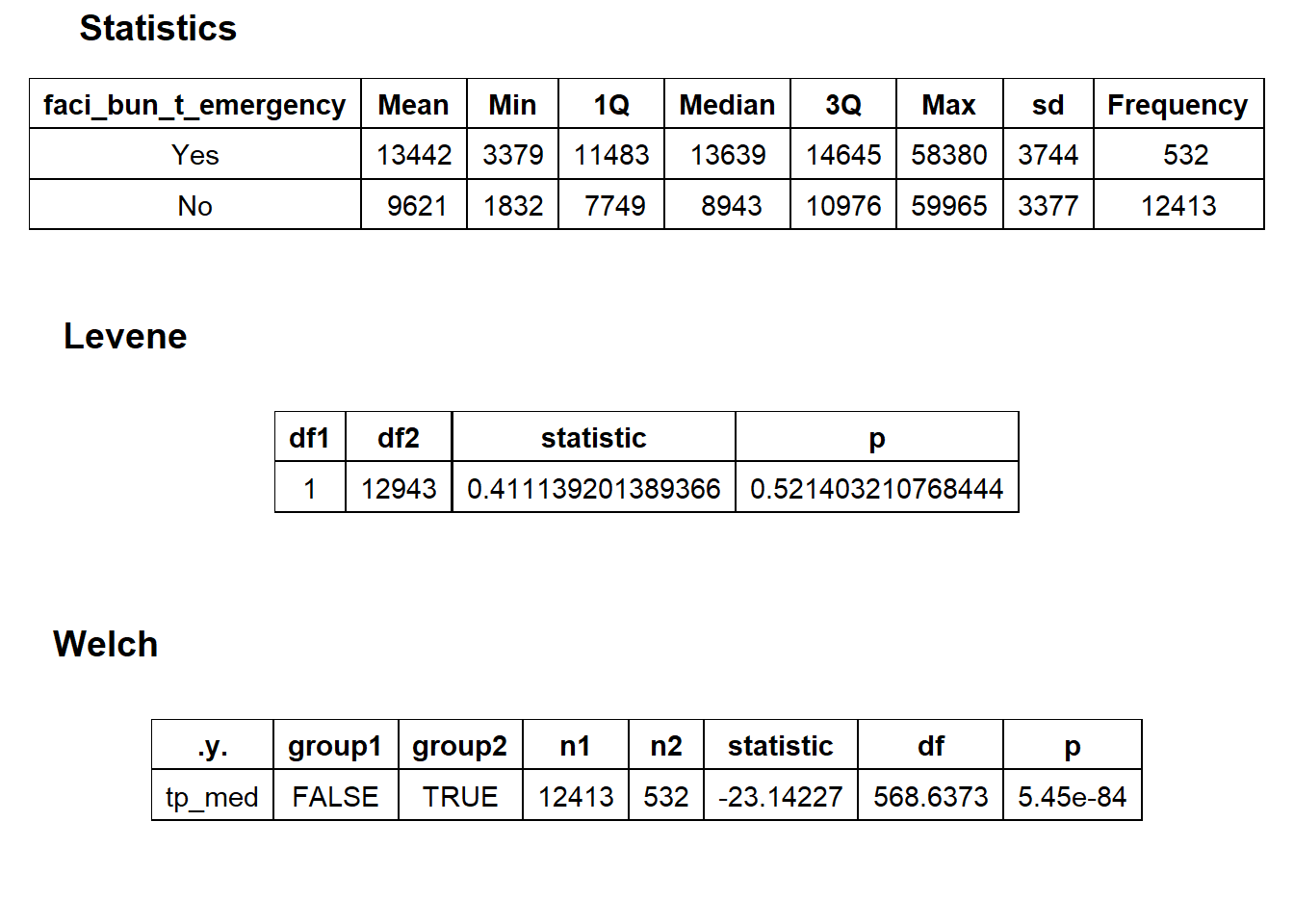
birth_vag_init_hosp_care <- birth_vag[faci_bun_t_initial==T & faci_bun_t_hospital==T & faci_bun_t_care==T]birth_vag <- birth_vag[faci_bun_t_initial==F & faci_bun_t_hospital==F & faci_bun_t_care==F]We will now examine the vaginal birth deliveries’s median price distribution and cost correlated tags.
birth_vag%>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.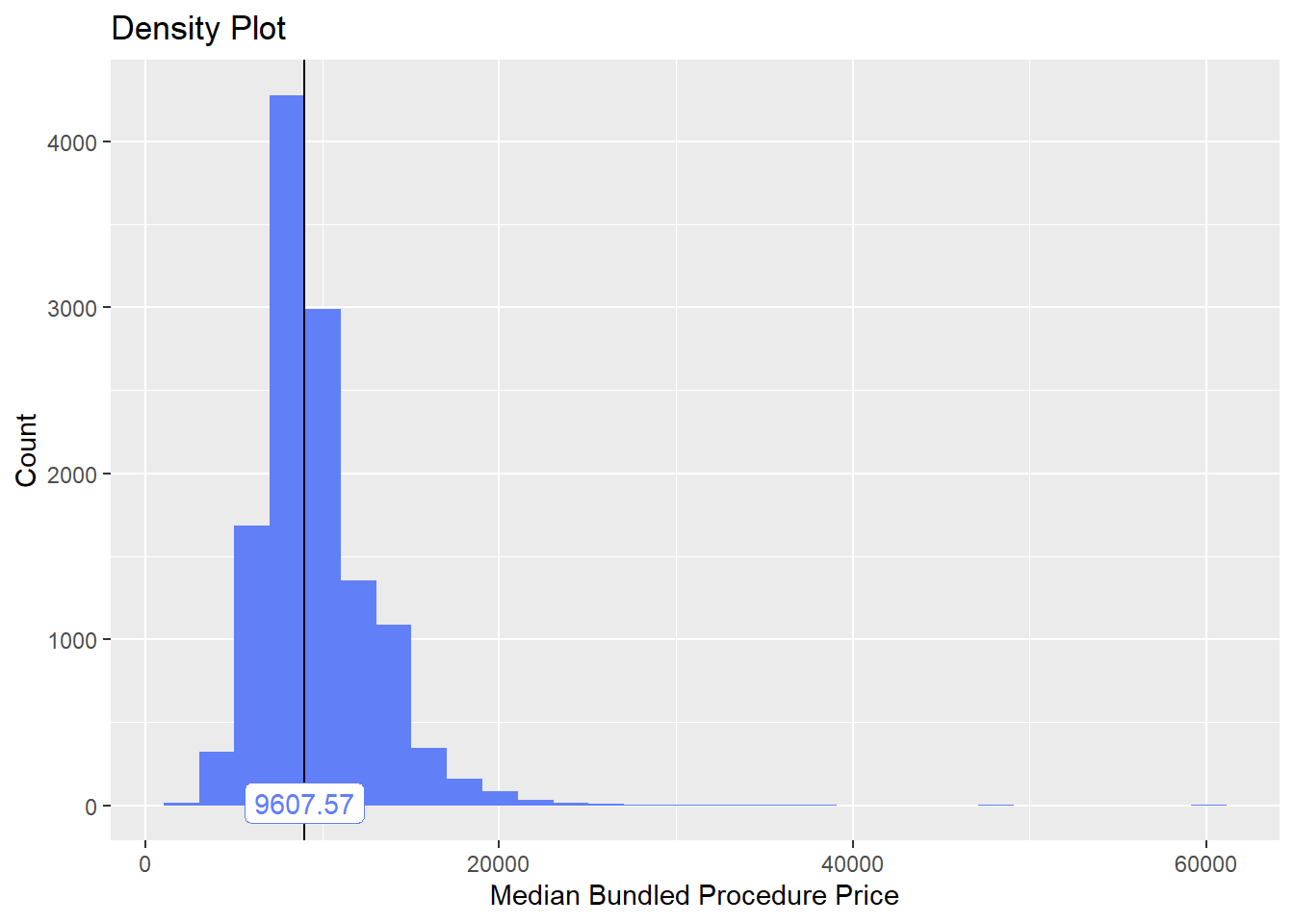
birth_vag%>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 276 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_anes 0.196
## 2 anes_bun_t_anesth 0.196
## 3 faci_bun_t_dept 0.192
## 4 faci_bun_t_emergency 0.192
## 5 duration_max 0.192
## 6 anes_bun_t_vag 0.191
## 7 anes_bun_t_deliver 0.190
## 8 anes_bun_t_liver 0.190
## 9 anes_bun_t_analg 0.188
## 10 faci_bun_t_visit 0.178
## # ... with 266 more rowsbirth_vag %>% get_tag_density_information("anes_bun_t_anes") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ anes_bun_t_anes"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 381## $dist_plots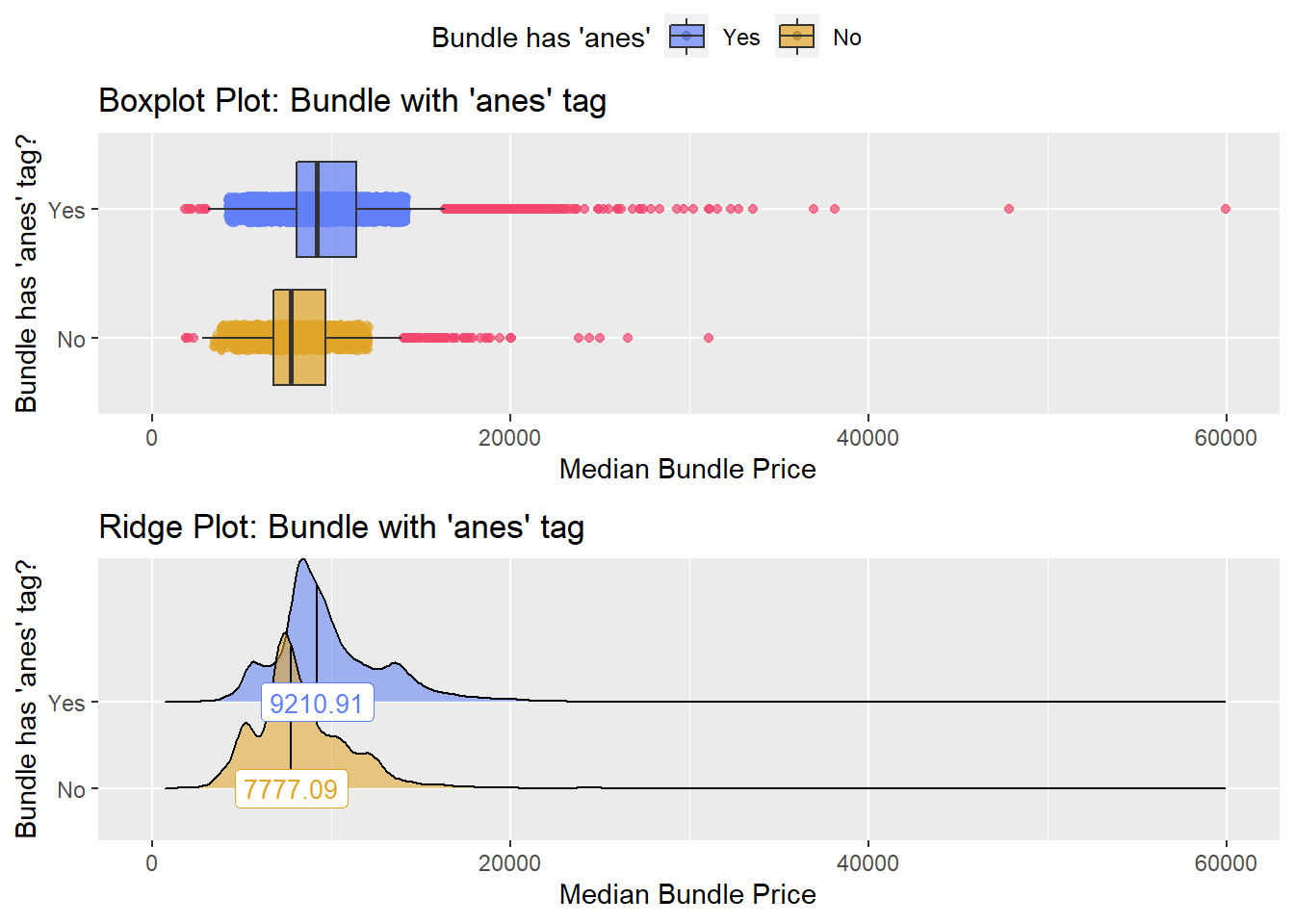
##
## $stat_tables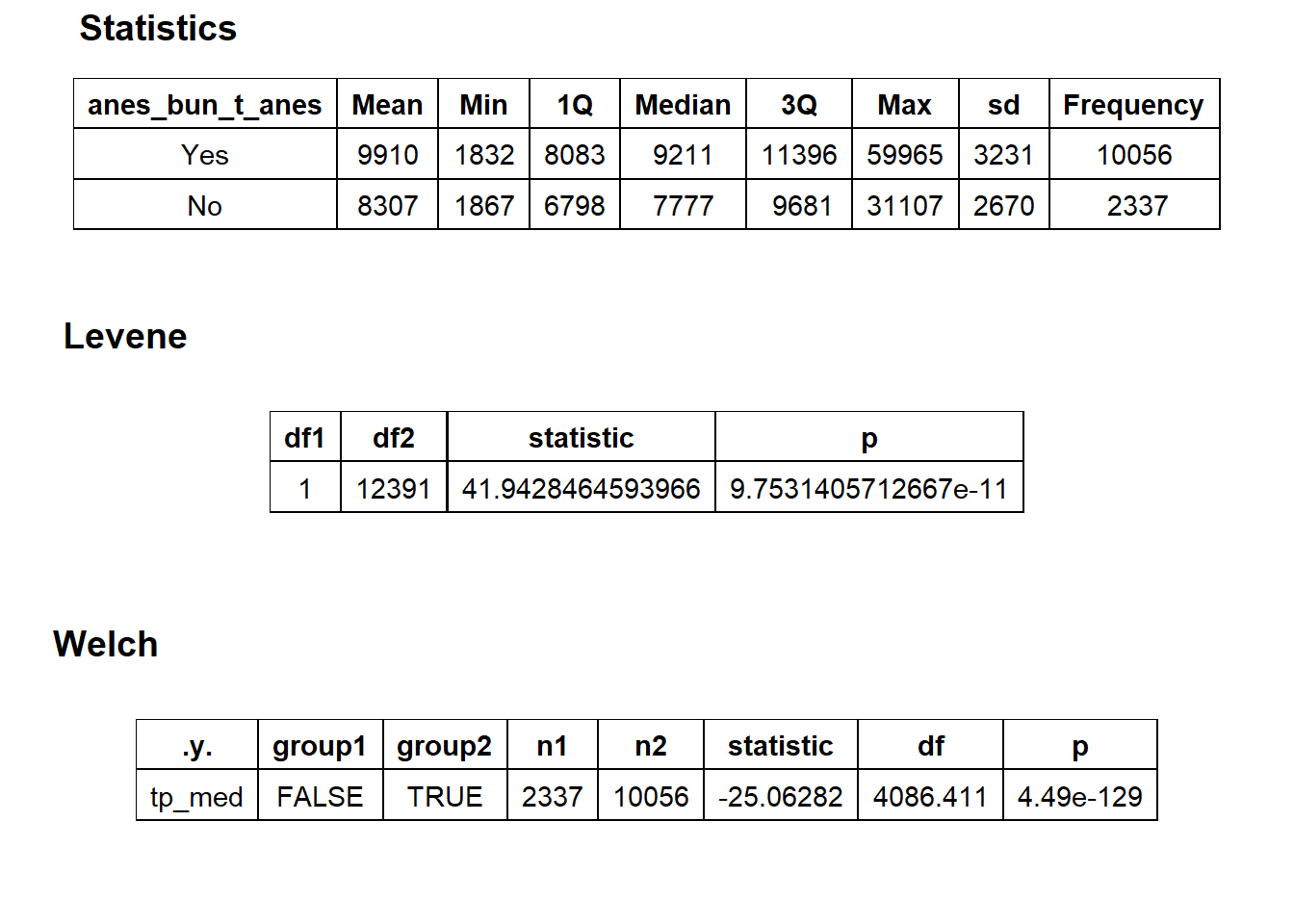
birth_vag <- birth_vag[anes_bun_t_anes==T]
birth_bag_no_anes <- birth_vag[anes_bun_t_anes==F]We will now examine the vaginal birth deliveries’s median price distribution and cost correlated tags.
birth_vag%>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
birth_vag%>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 259 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_dept 0.206
## 2 faci_bun_t_emergency 0.206
## 3 faci_bun_t_visit 0.186
## 4 duration_max 0.178
## 5 surg_bun_t_hyst 0.144
## 6 surg_bun_t_hysterectomy 0.144
## 7 path_bun_t_patho 0.127
## 8 path_bun_t_pathologist 0.127
## 9 path_bun_t_tissue 0.127
## 10 surg_bun_t_total 0.117
## # ... with 249 more rowsbirth_vag %>% get_tag_density_information("faci_bun_t_emergency") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ faci_bun_t_emergency"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 331## $dist_plots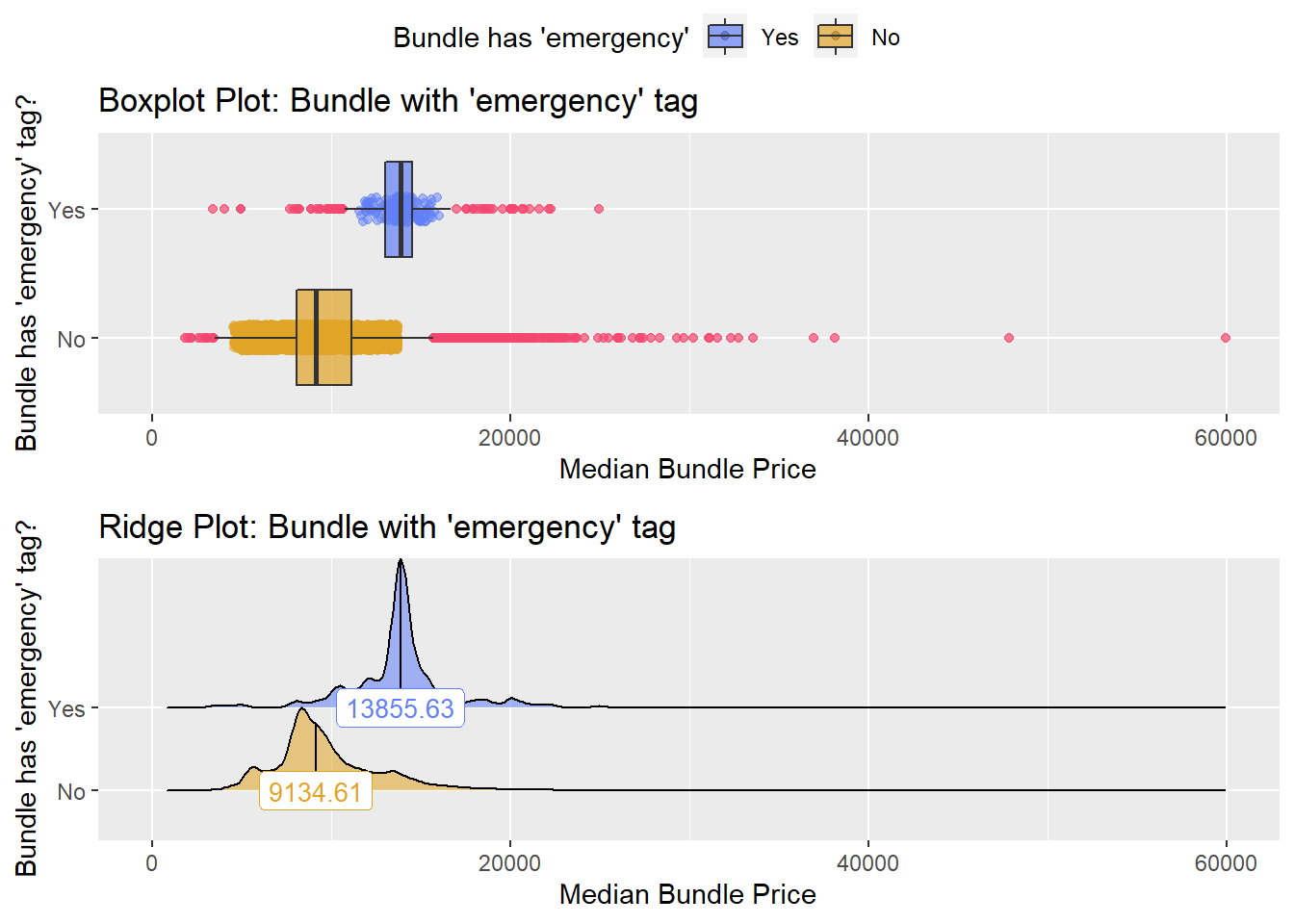
##
## $stat_tables
birth_vag_w_anes_w_emerg_dept <- birth_vag[faci_bun_t_emergency==T & faci_bun_t_dept==T]
birth_vag <- birth_vag[faci_bun_t_emergency==F & faci_bun_t_dept==F]We will now examine the vaginal birth deliveries’s median price distribution and cost correlated tags.
birth_vag%>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.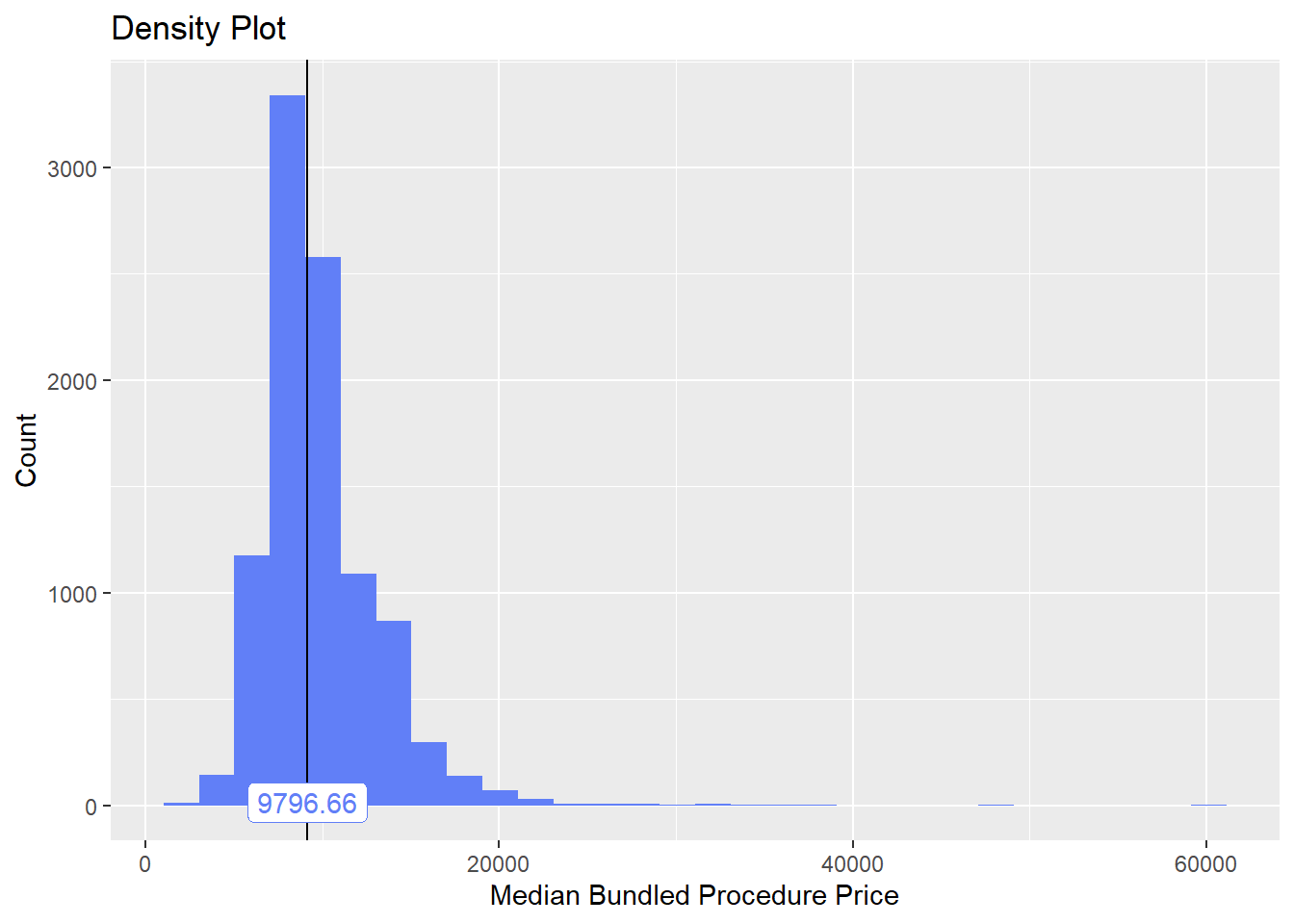
birth_vag%>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 241 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.179
## 2 surg_bun_t_hyst 0.150
## 3 surg_bun_t_hysterectomy 0.150
## 4 surg_bun_t_total 0.121
## 5 path_bun_t_patho 0.117
## 6 path_bun_t_pathologist 0.117
## 7 path_bun_t_tissue 0.117
## 8 anes_bun_t_lower 0.115
## 9 anes_bun_t_abdom 0.108
## 10 anes_bun_t_surg 0.0929
## # ... with 231 more rowsbirth_vag %>% get_tag_density_information("surg_bun_t_hysterectomy") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_hysterectomy"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 1590## $dist_plots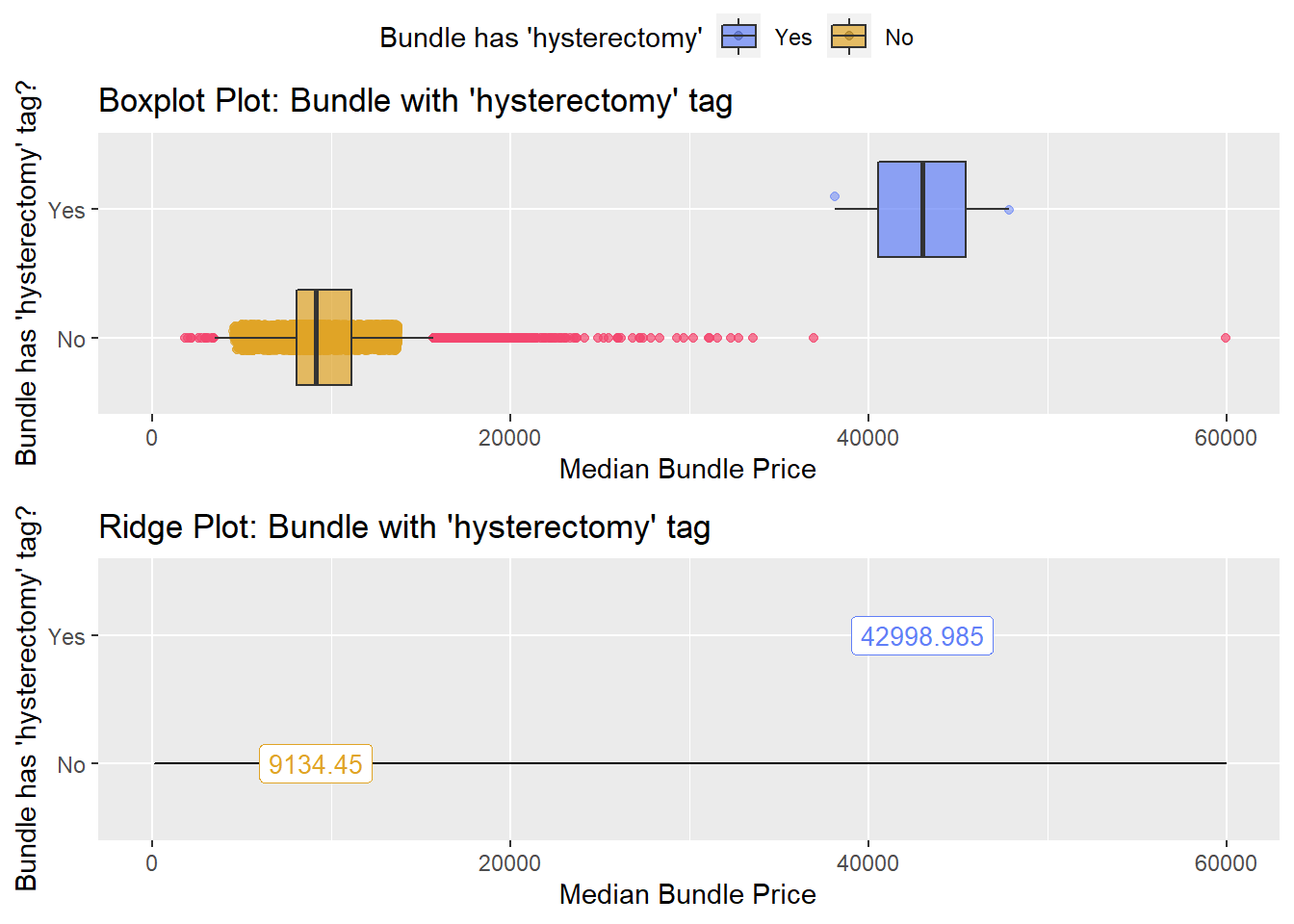
##
## $stat_tables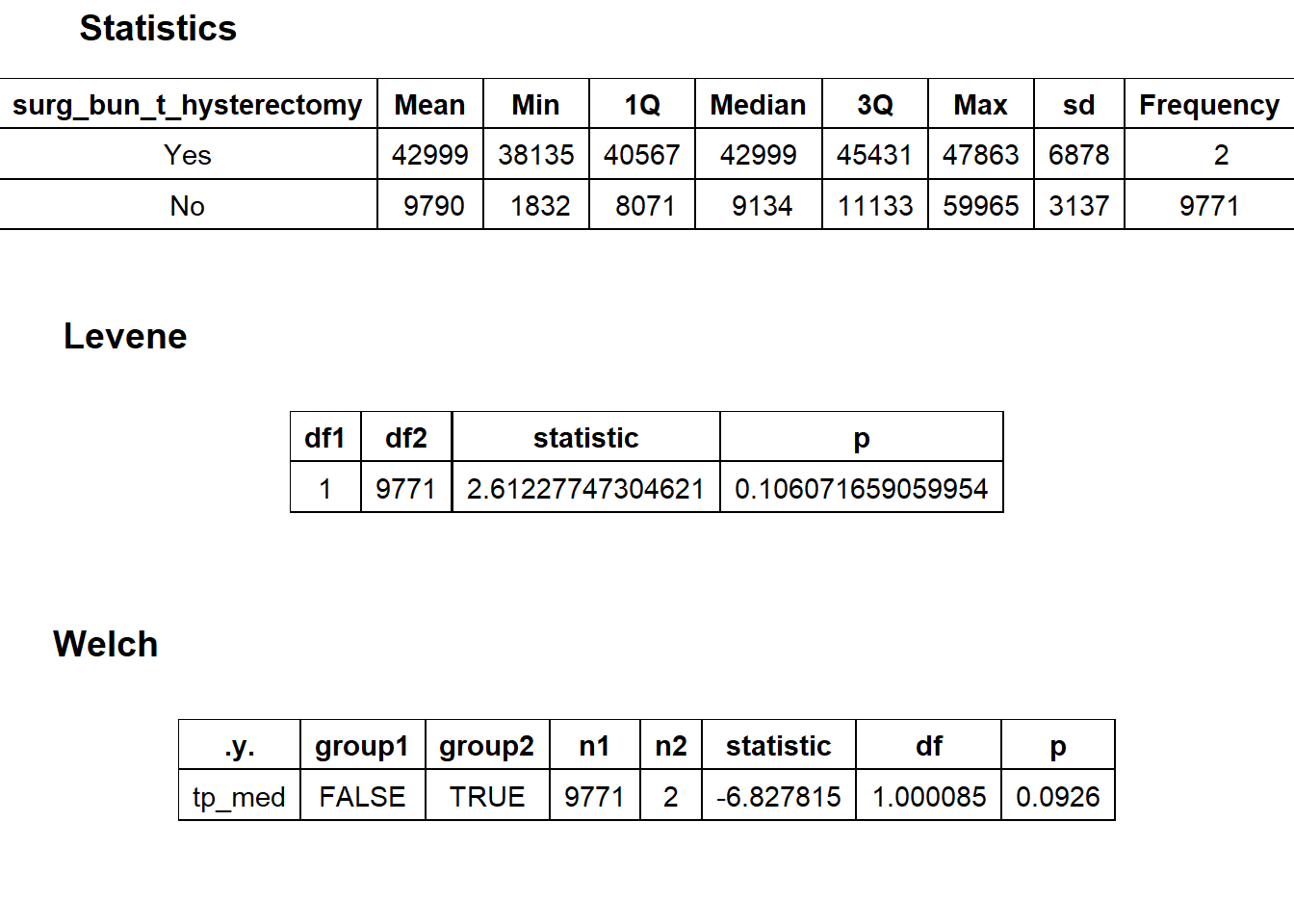
birth_vag <- birth_vag[surg_bun_t_hysterectomy==F]birth_vag %>% get_tag_density_information("path_bun_t_tissue") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_tissue"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 641## $dist_plots
##
## $stat_tables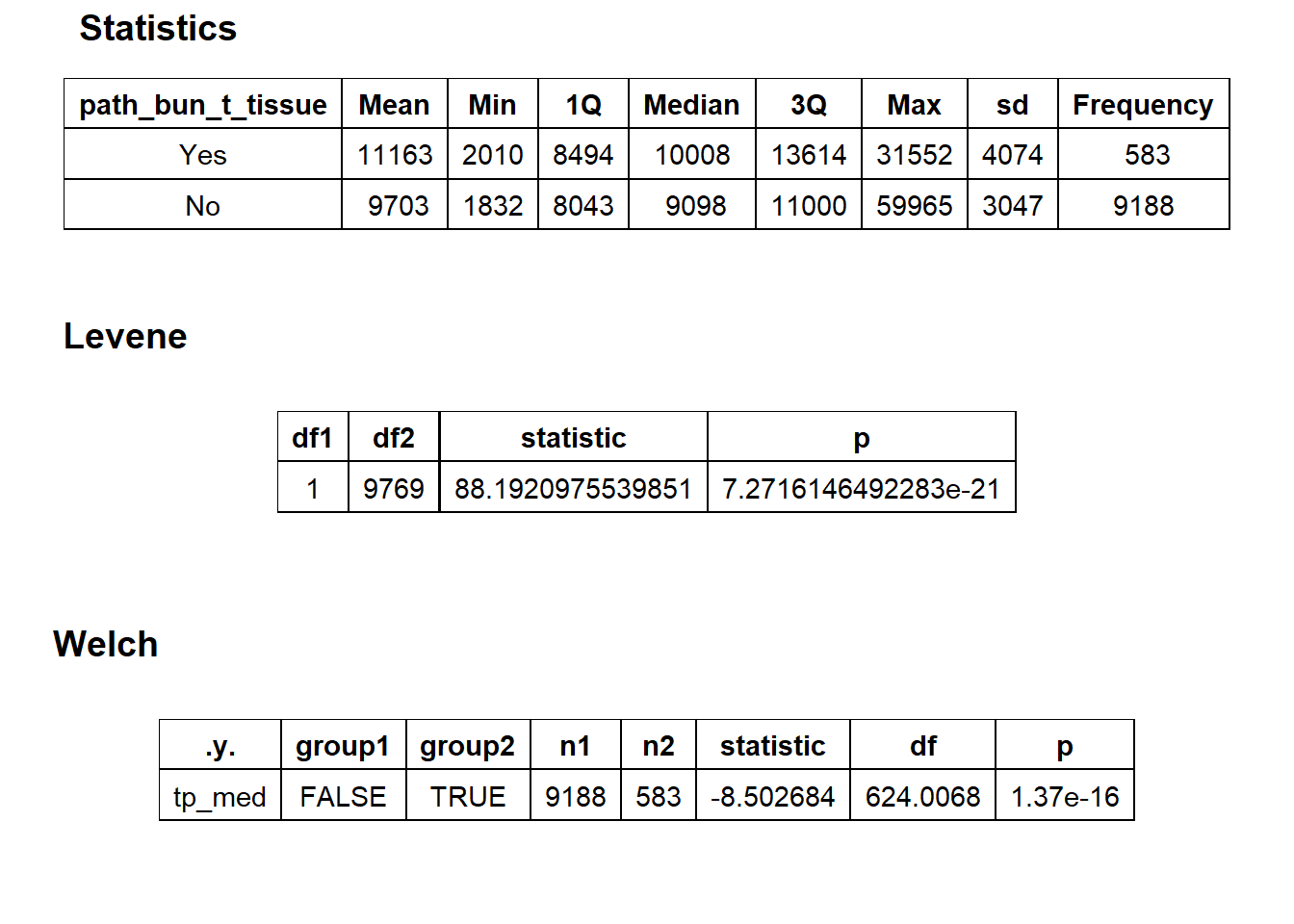
We will now examine the vaginal birth deliveries’s median price distribution and cost correlated tags.
birth_vag%>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
birth_vag%>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 237 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.178
## 2 path_bun_t_patho 0.110
## 3 path_bun_t_pathologist 0.110
## 4 path_bun_t_tissue 0.110
## 5 path_bun_t_complete 0.0754
## 6 surg_bun_t_uterus 0.0741
## 7 path_bun_t_screen 0.0732
## 8 anes_bun_t_tubal 0.0711
## 9 surg_bun_t_fallopian 0.0709
## 10 surg_bun_t_tube 0.0709
## # ... with 227 more rowsbirth_vag %>% get_tag_density_information("path_bun_t_patho") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_patho"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 641## $dist_plots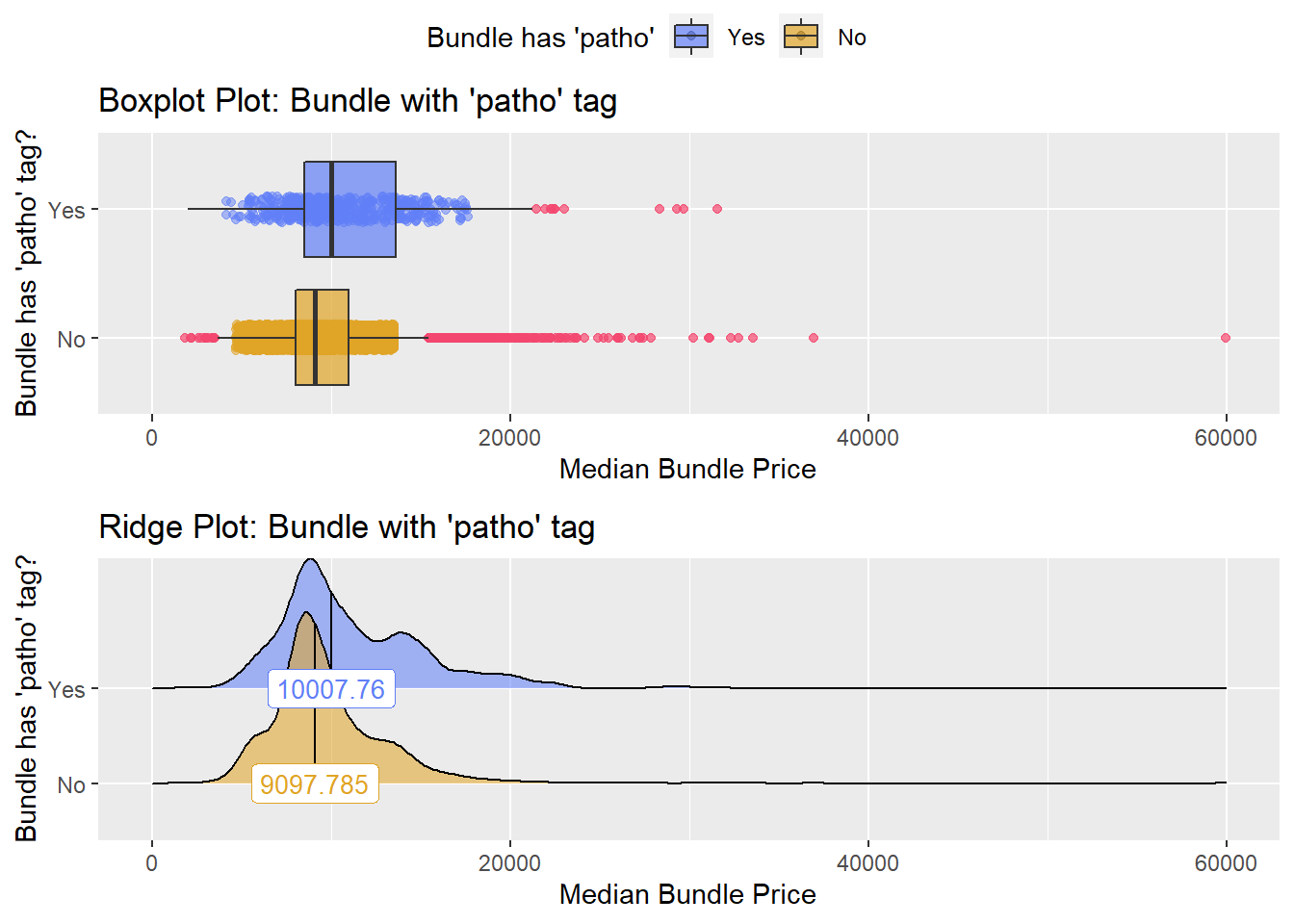
##
## $stat_tables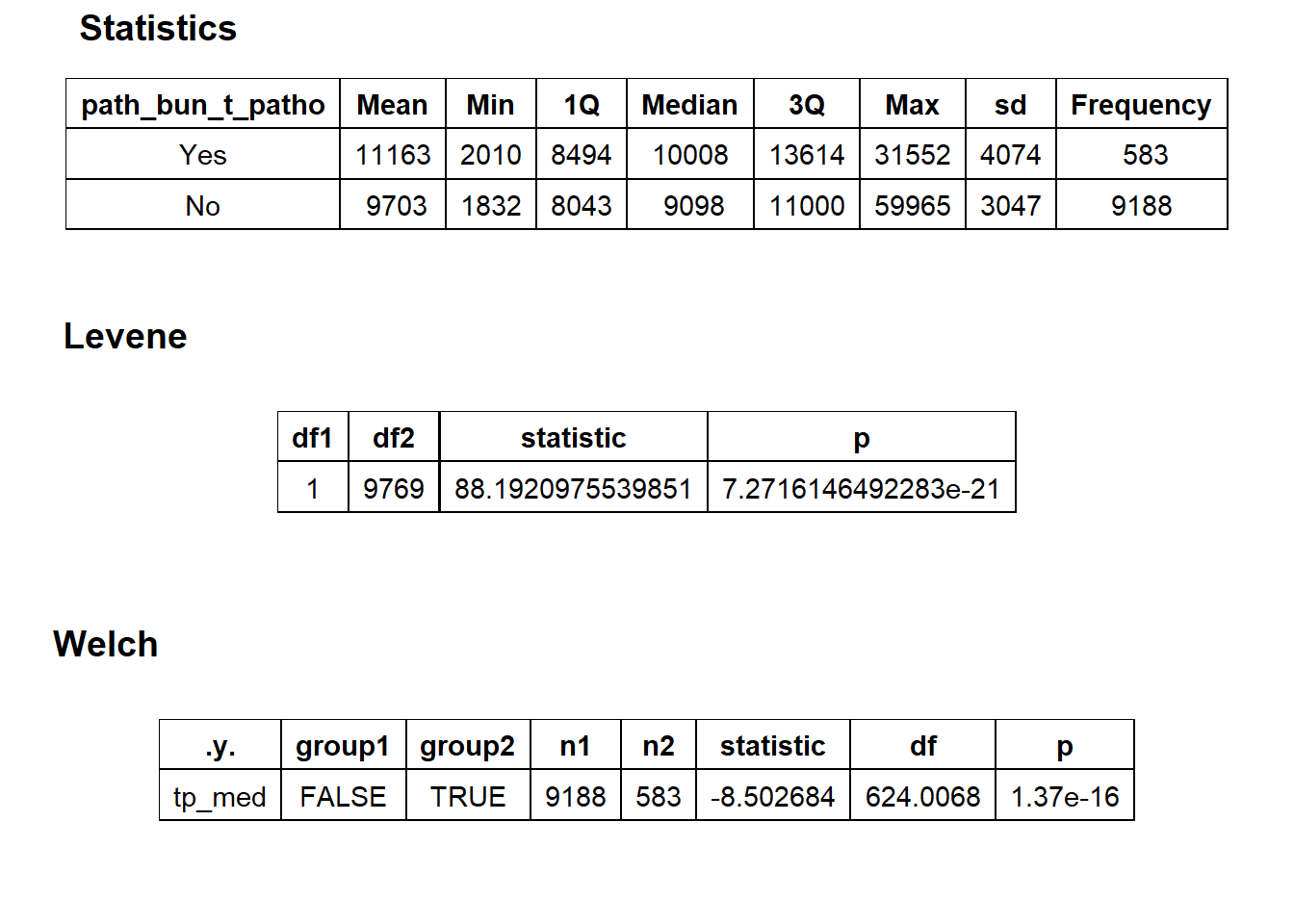
birth_vag_with_patho <- birth_vag[path_bun_t_patho==T]
birth_vag <- birth_vag[path_bun_t_patho==F]We will now examine the vaginal birth deliveries’s median price distribution and cost correlated tags.
birth_vag %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
birth_vag %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 211 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.168
## 2 surg_bun_t_uterus 0.079
## 3 path_bun_t_complete 0.0723
## 4 path_bun_t_screen 0.0713
## 5 surg_bun_t_hematoma 0.0703
## 6 radi_bun_t_ct 0.0612
## 7 medi_bun_t_observ 0.061
## 8 anes_bun_t_abdom 0.0597
## 9 anes_bun_t_lower 0.0597
## 10 radi_bun_t_angiography 0.0594
## # ... with 201 more rowsbirth_vag[surg_bun_t_uterus==T] %>% nrow()## [1] 1birth_vag %>% get_tag_density_information("path_bun_t_complete") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_complete"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 509## $dist_plots
##
## $stat_tables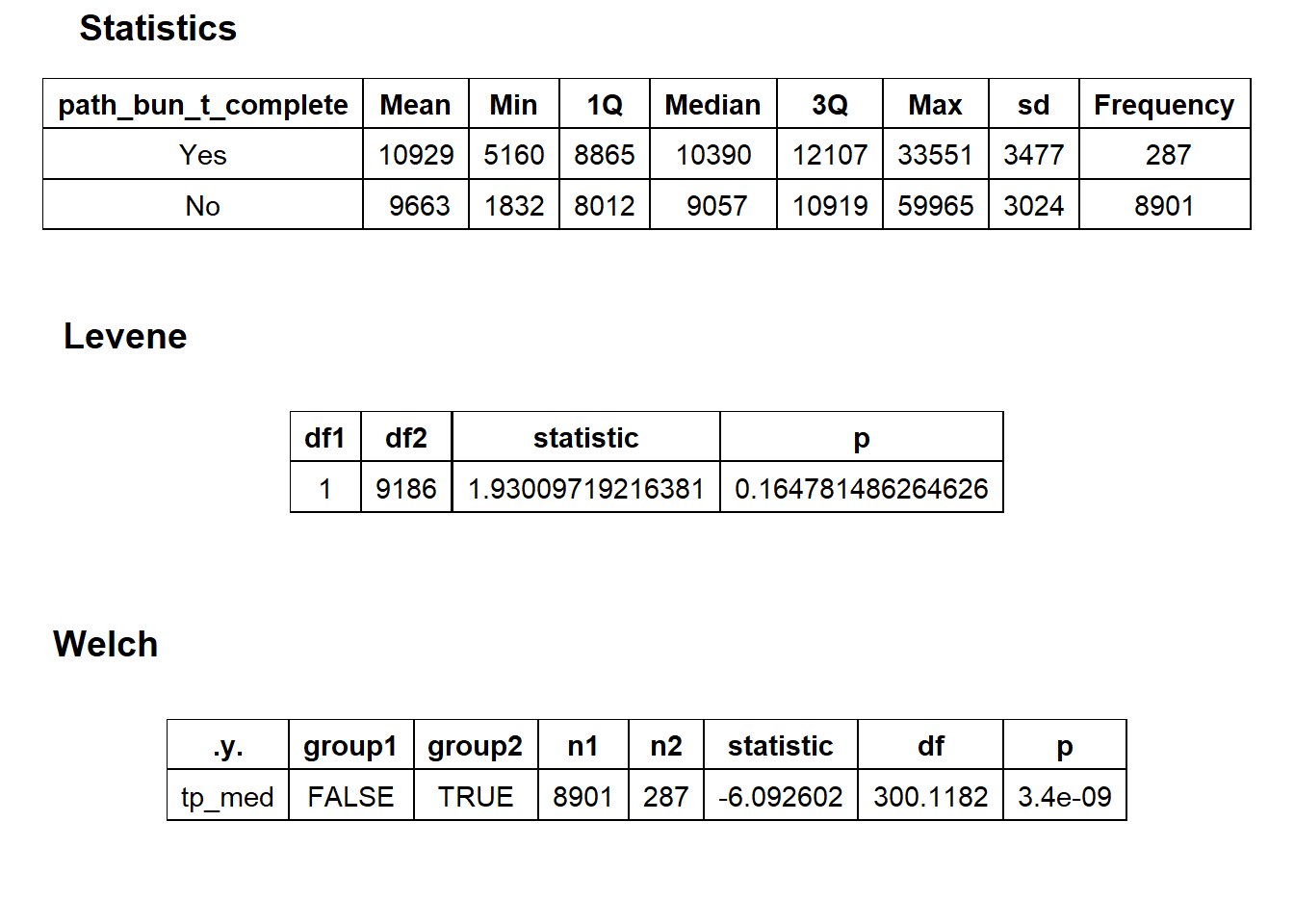
birth_vag %>% get_tag_density_information("radi_bun_t_ct") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.
## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ radi_bun_t_ct"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 2310## $dist_plots
##
## $stat_tables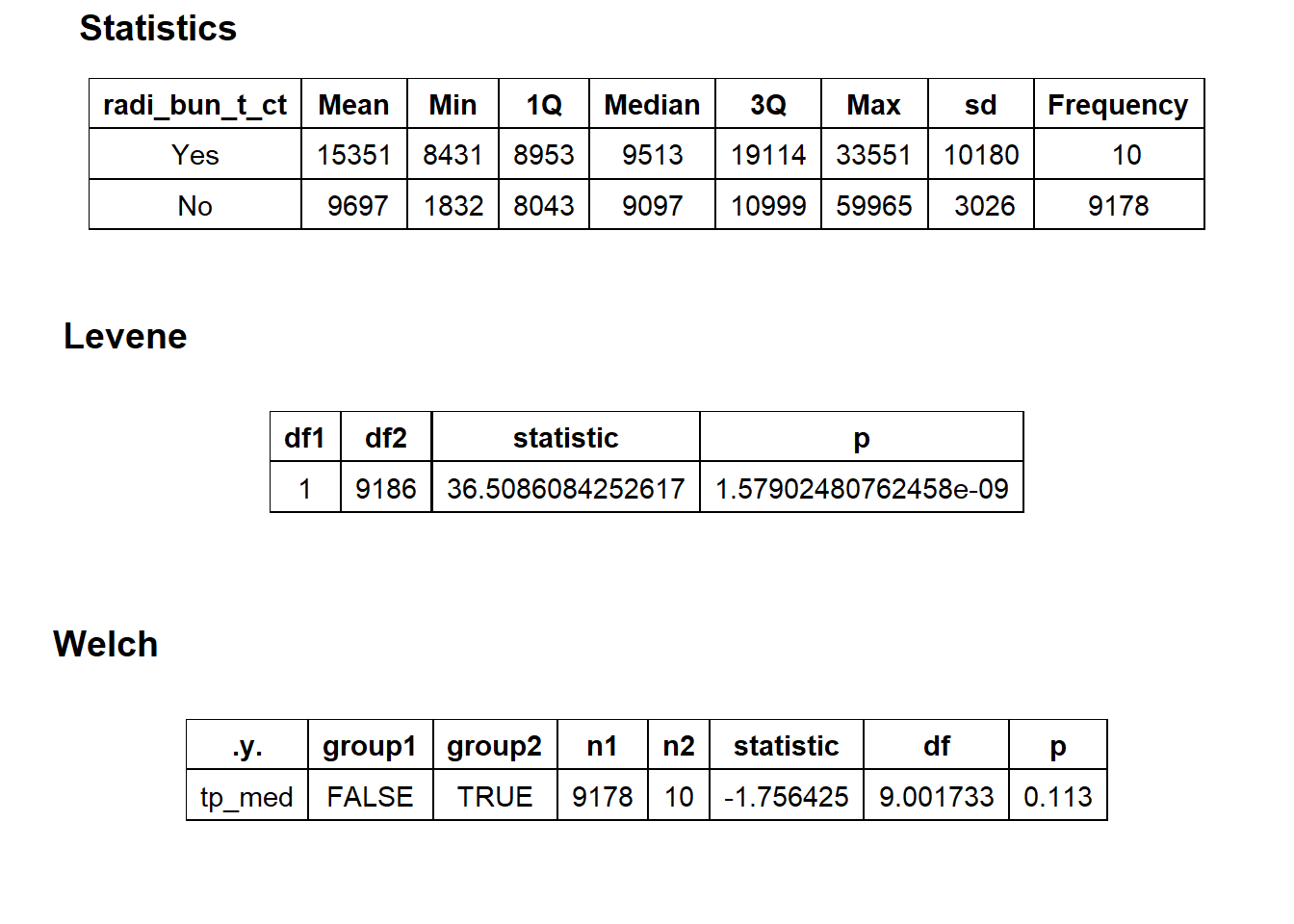
birth_vag <- birth_vag[surg_bun_t_uterus==F & path_bun_t_complete==F & radi_bun_t_ct==F]birth_vag %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
birth_vag %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 186 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.159
## 2 path_bun_t_anti 0.0597
## 3 path_bun_t_antibod 0.0597
## 4 path_bun_t_screen 0.0558
## 5 radi_bun_t_pelvi 0.0548
## 6 surg_bun_t_dev 0.0522
## 7 surg_bun_t_device 0.0522
## 8 medi_bun_t_observ 0.0514
## 9 surg_bun_t_insert 0.0476
## 10 path_bun_t_blood 0.0468
## # ... with 176 more rowsbirth_vag[cnt > 10 & faci_bun_sum_med > 2000] %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.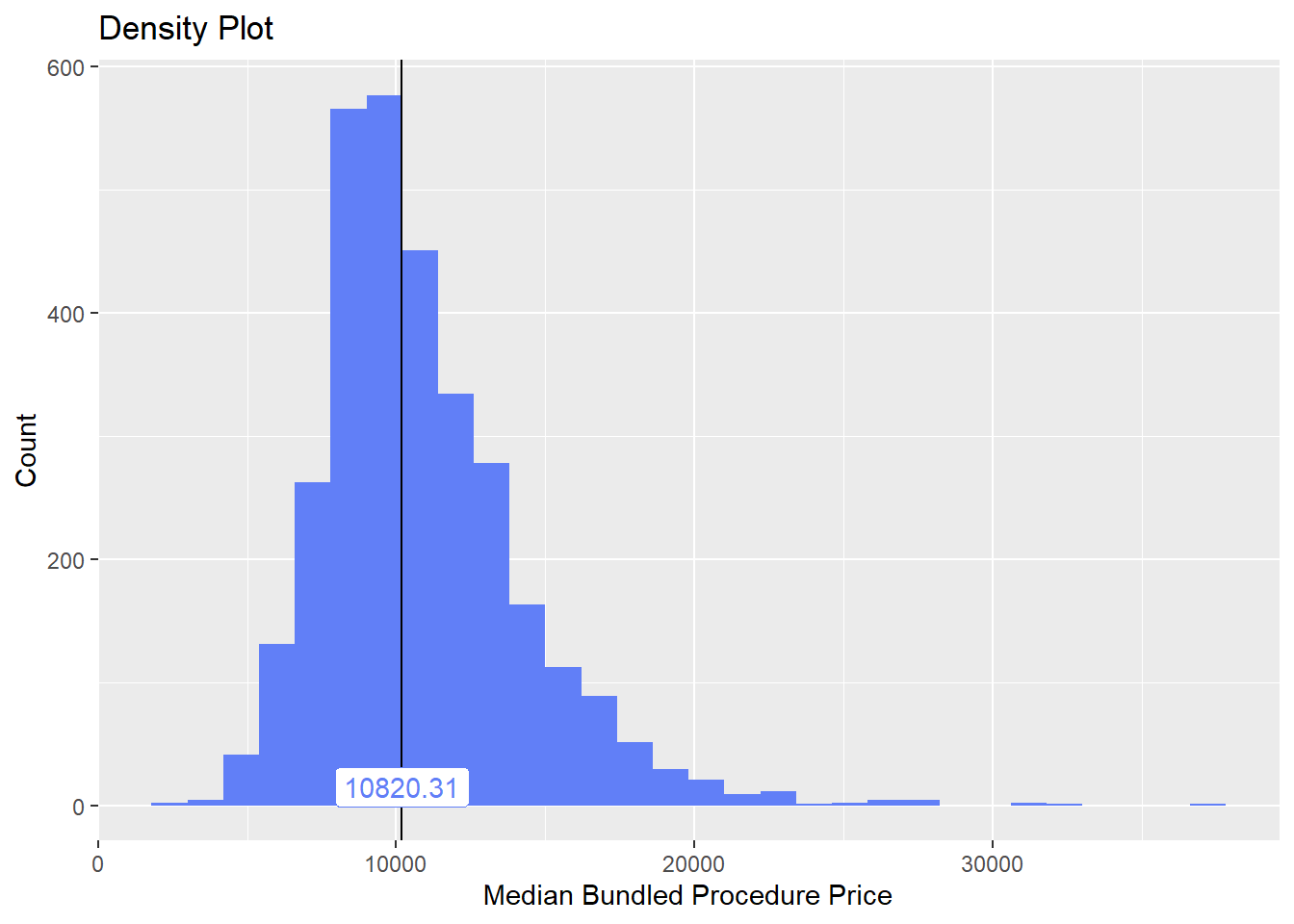
birth_vag[cnt > 10 & faci_bun_sum_med > 2000] %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 171 x 2
## name correlation
## <chr> <dbl>
## 1 medi_bun_t_clinic 0.123
## 2 medi_bun_t_outpt 0.123
## 3 duration_max 0.122
## 4 medi_bun_t_visit 0.116
## 5 radi_bun_t_pelvi 0.086
## 6 path_bun_t_anti 0.0721
## 7 path_bun_t_antibod 0.0721
## 8 medi_bun_t_inject 0.0641
## 9 medi_bun_t_sulfate 0.0629
## 10 surg_bun_t_remov 0.0618
## # ... with 161 more rowsbirth_vag[cnt > 10 & faci_bun_sum_med > 2000] %>% nrow()## [1] 3146birth_vag <- birth_vag[cnt > 10 & faci_bun_sum_med > 2000]birth_vag %>% get_tag_density_information("medi_bun_t_clinic") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_clinic"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 513## $dist_plots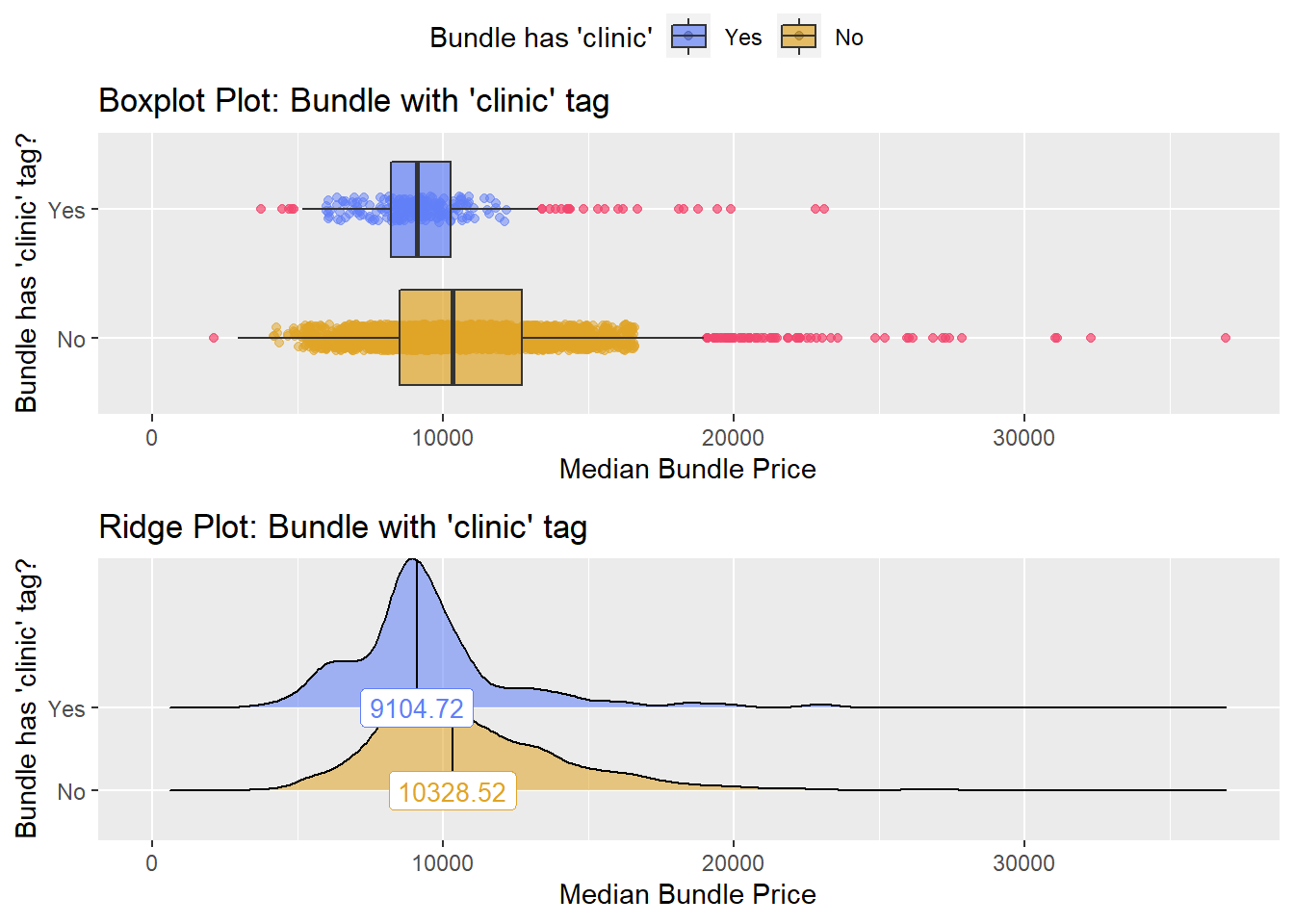
##
## $stat_tables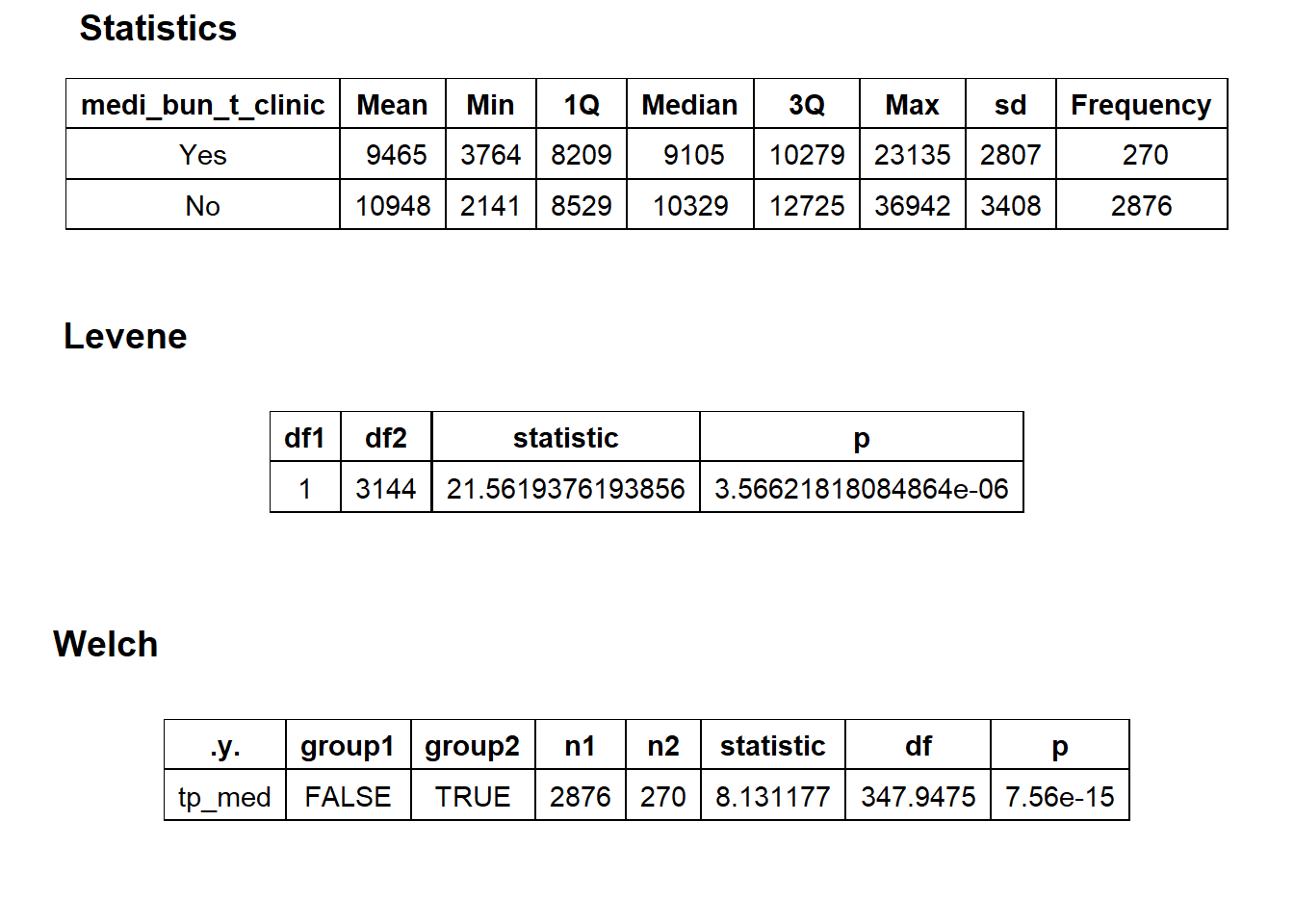
birth_vag <- birth_vag[medi_bun_t_clinic==F]birth_vag %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
birth_vag %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 167 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.119
## 2 radi_bun_t_pelvi 0.0882
## 3 path_bun_t_fetal 0.0782
## 4 path_bun_t_anti 0.0756
## 5 path_bun_t_antibod 0.0756
## 6 path_bun_t_typing 0.073
## 7 path_bun_t_blood 0.0631
## 8 surg_bun_t_remov 0.0623
## 9 surg_bun_t_removal 0.0554
## 10 anes_bun_t_vag 0.0543
## # ... with 157 more rowsbirth_vag %>% get_tag_density_information("radi_bun_t_pelvi") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ radi_bun_t_pelvi"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 3920## $dist_plots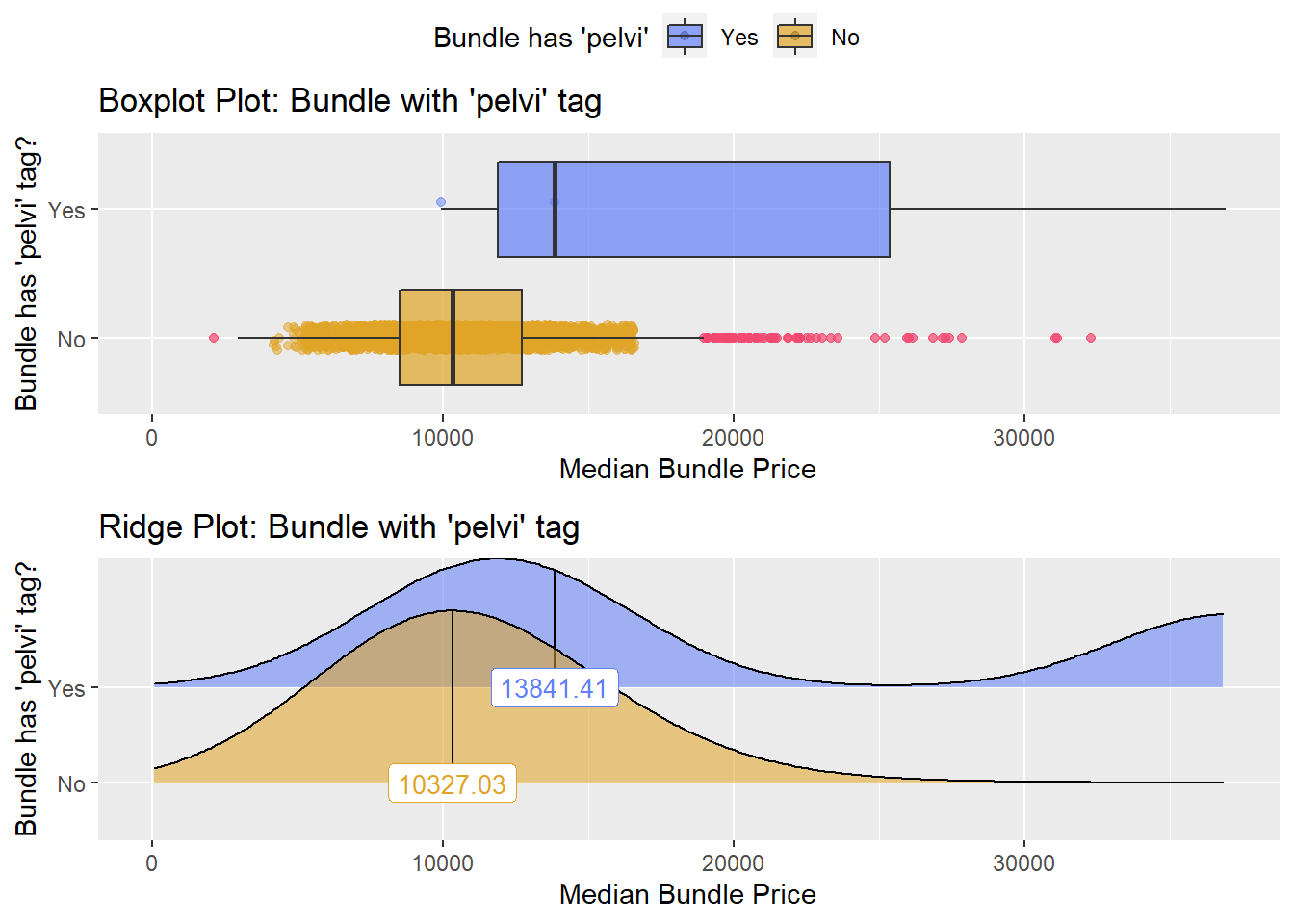
##
## $stat_tables
birth_vag <- birth_vag[radi_bun_t_pelvi==F]birth_vag %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.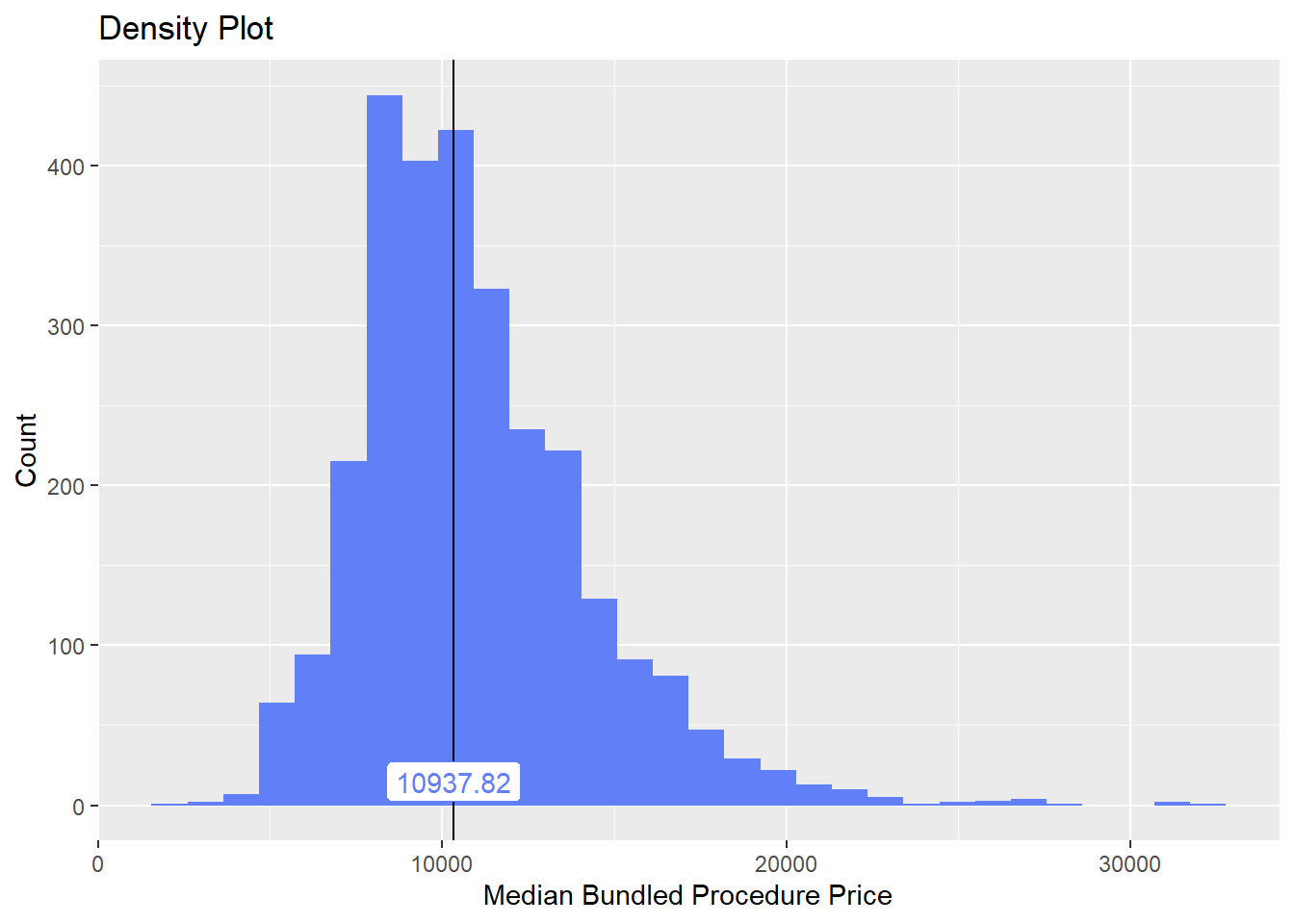
birth_vag %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 166 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.117
## 2 path_bun_t_fetal 0.079
## 3 path_bun_t_anti 0.0765
## 4 path_bun_t_antibod 0.0765
## 5 path_bun_t_typing 0.0739
## 6 path_bun_t_blood 0.0639
## 7 surg_bun_t_remov 0.0631
## 8 surg_bun_t_removal 0.0561
## 9 anes_bun_t_vag 0.055
## 10 surg_bun_t_scar 0.0543
## # ... with 156 more rowsbirth_vag[path_bun_t_fetal==T] %>% nrow()## [1] 1birth_vag <- birth_vag[path_bun_t_fetal==F]birth_vag %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
birth_vag %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 165 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.114
## 2 surg_bun_t_remov 0.0634
## 3 surg_bun_t_removal 0.0564
## 4 anes_bun_t_vag 0.0553
## 5 surg_bun_t_scar 0.0545
## 6 surg_bun_t_treat 0.0545
## 7 anes_bun_t_analg 0.054
## 8 surg_bun_t_fallopian 0.0533
## 9 surg_bun_t_tube 0.0533
## 10 anes_bun_t_tubal 0.0533
## # ... with 155 more rowsbtb for vaginal births
birth_vag_btbv4 <- birth_vag %>% btbv4()birth_vag_bq <- birth_vag_btbv4[,
primary_doctor := pmap(.l=list(doctor_npi1=doctor_npi_str1,
doctor_npi2=doctor_npi_str2,
class_reqs="Obstetrics & Gynecology"#,
# specialization_reqs = "Obstetrics|||Maternal & Fetal Medicine"
),
.f=calculate_primary_doctor) %>% as.character()
] %>%
#Filter out any procedures where our doctors fail both criteria.
.[!(primary_doctor %in% c("BOTH_DOC_FAIL_CRIT", "TWO_FIT_ALL_SPECS"))] %>%
.[,primary_doctor_npi := fifelse(primary_doctor==doctor_str1,
doctor_npi_str1,
doctor_npi_str2)] %>%
.[,`:=`(procedure_type=5, procedure_modifier="Vaginal Delivery")]## [1] "multiple meet class req"
## [1] "TIFFANY KRISTEN WEBER" "HEATHER DONAHOE CAMPBELL"
## [1] "multiple meet class req"
## [1] "BRYAN SETH PALMER" "KIRK ANDREW LAMMI"
## [1] "multiple meet class req"
## [1] "DAVID WARE BRANCH" "STEVEN M THACKERAY"
## [1] "multiple meet class req"
## [1] "STEVEN GREG NANCE" "DONNA DIZON-TOWNSON"
## [1] "multiple meet class req"
## [1] "LUNT KVARFORDT (SAINT GEORGE)" "CHAD C LUNT"
## [1] "multiple meet class req"
## [1] "DAVID WARE BRANCH" "ANN BRUNO"
## [1] "multiple meet class req"
## [1] "HANNELE MARIE LAINE" "TROY FLINT PORTER"
## [1] "multiple meet class req"
## [1] "LUNT KVARFORDT (SAINT GEORGE)" "TRACY D KVARFORDT"
## [1] "multiple meet class req"
## [1] "KAREN BOHEEN (LAYTON)" "KAREN BOHEEN"
## [1] "multiple meet class req"
## [1] "JEFF DAVID NANCE" "STEVEN GREG NANCE"
## [1] "multiple meet class req"
## [1] "KAREN BOHEEN (LAYTON)" "KAREN BOHEEN"
## [1] "multiple meet class req"
## [1] "TROY FLINT PORTER" "CARA C HEUSER"
## [1] "multiple meet class req"
## [1] "JEFFOREY R. THORPE" "PETER GILSON DREWES"
## [1] "multiple meet class req"
## [1] "DAVID E LUDLOW" "JASON KEITH MORRIS"
## [1] "multiple meet class req"
## [1] "ROBERT MERRILL" "JEFFREY DENNIS QUINN"
## [1] "multiple meet class req"
## [1] "DAVID WARE BRANCH" "LISA MARIE GRAVELLE"
## [1] "multiple meet class req"
## [1] "JEFFREY CHARLES BROBERG" "BRIAN L WOLSEY"
## [1] "multiple meet class req"
## [1] "MARIA A ONEIDA" "MICHAEL S ESPLIN"
## [1] "multiple meet class req"
## [1] "DAVID WARE BRANCH" "JANE C BOWMAN"
## [1] "multiple meet class req"
## [1] "MARCELA SMID" "AMY E SULLIVAN"
## [1] "multiple meet class req"
## [1] "DAVID KEITH TUROK" "AMY E SULLIVAN"
## [1] "multiple meet class req"
## [1] "JEFF DAVID NANCE" "DONNA DIZON-TOWNSON"
## [1] "multiple meet class req"
## [1] "SCOTT R. JACOB" "PETER GILSON DREWES"
## [1] "multiple meet class req"
## [1] "CLAYTON SCOTT SYNDERGAARD" "BEN DOUGLAS WILLIAMS"
## [1] "multiple meet class req"
## [1] "CRAIG L HURST MD (LAYTON)" "CRAIG L HURST"
## [1] "multiple meet class req"
## [1] "ERIN A.S. CLARK" "ANN BRUNO"
## [1] "multiple meet class req"
## [1] "IBRAHIM A HAMMAD" "AMY E SULLIVAN"
## [1] "multiple meet class req"
## [1] "DAVID BIERER" "ROSEMARY TIRINNANZI LESSER"
## [1] "multiple meet class req"
## [1] "JAMES P LAMOREAUX" "SEAN HASKETT"
## [1] "multiple meet class req"
## [1] "JULIE GLENN GROVER" "DONNA DIZON-TOWNSON"
## [1] "multiple meet class req"
## [1] "STEVEN GREG NANCE" "HELEN FELTOVICH"
## [1] "multiple meet class req"
## [1] "CHRISTIAN D FROERER" "JEFFREY H BARTON"
## [1] "multiple meet class req"
## [1] "SHANNON L TILLY" "NANCY C ROSE"
## [1] "multiple meet class req"
## [1] "JASON KEITH MORRIS" "JEANETTE RUTH CARPENTER"
## [1] "multiple meet class req"
## [1] "STEVEN GREG NANCE" "GLENN K SCHEMMER"
## [1] "multiple meet class req"
## [1] "COURTNEY CLAIRE MACLEAN" "ROBERT M. SILVER"
## [1] "multiple meet class req"
## [1] "SHANNON L TILLY" "HANNELE MARIE LAINE"
## [1] "multiple meet class req"
## [1] "ALICIA TABISH JONES" "DOUGLAS S RICHARDS"
## [1] "multiple meet class req"
## [1] "ASHLEY CRAMER BYNO" "HANNELE MARIE LAINE"
## [1] "multiple meet class req"
## [1] "TENNILLE CLOWARD" "DONNA DIZON-TOWNSON"
## [1] "multiple meet class req"
## [1] "TRACY W WINWARD" "BRADY BENHAM"
## [1] "multiple meet class req"
## [1] "DAVID A KIRKMAN" "ANNE SADLER BLACKETT"
## [1] "multiple meet class req"
## [1] "JENNA MADSEN" "KRISTIN ELIZABETH WEXLER"
## [1] "multiple meet class req"
## [1] "DAVID B YOUNG" "JEANETTE RUTH CARPENTER"
## [1] "multiple meet class req"
## [1] "HOWARD T. SHARP" "LAUREN H THEILEN"
## [1] "multiple meet class req"
## [1] "BRANDON LEE REYNOLDS" "ROBERT M. SILVER"
## [1] "multiple meet class req"
## [1] "JHENETTE RENEE LAUDER" "MICHELLE LYNNE PRECOURT DEBBINK"
## [1] "multiple meet class req"
## [1] "EVAN JONES" "SEAN HASKETT"
## [1] "multiple meet class req"
## [1] "MARGIT SZABO LISTER" "MARGRET ALLISON MENCER"
## [1] "multiple meet class req"
## [1] "DAVID WARE BRANCH" "MICHAEL L DRAPER"
## [1] "multiple meet class req"
## [1] "JEFFREY CHARLES BROBERG" "JULIE GAINER"
## [1] "multiple meet class req"
## [1] "MELISSA ANN BROWN" "TROY FLINT PORTER"
## [1] "multiple meet class req"
## [1] "SHANNON L TILLY" "DOUGLAS S RICHARDS"
## [1] "multiple meet class req"
## [1] "ANDREA JULIET HEBERT" "TROY FLINT PORTER"
## [1] "multiple meet class req"
## [1] "CHRISTIAN D FROERER" "CARA C HEUSER"
## [1] "multiple meet class req"
## [1] "JULIE GLENN GROVER" "GLENN K SCHEMMER"
## [1] "multiple meet class req"
## [1] "JEFF DAVID NANCE" "HELEN FELTOVICH"
## [1] "multiple meet class req"
## [1] "DAVID WARE BRANCH" "TROY FLINT PORTER"
## [1] "multiple meet class req"
## [1] "ALEXANDRA GROSVENOR ELLER" "DOUGLAS S RICHARDS"
## [1] "multiple meet class req"
## [1] "JEFFREY CHARLES BROBERG" "GLENN K SCHEMMER"
## [1] "multiple meet class req"
## [1] "MICHAEL S ESPLIN" "NATALIE K LOEWEN"
## [1] "multiple meet class req"
## [1] "EVAN JONES" "RYAN C. OLLERTON"
## [1] "multiple meet class req"
## [1] "REBECCA LOUISE PONDER" "DOUGLAS S RICHARDS"
## [1] "multiple meet class req"
## [1] "SEAN HASKETT" "TENNILLE CLOWARD"
## [1] "multiple meet class req"
## [1] "ALEXANDRA GROSVENOR ELLER" "CHRISTIAN D FROERER"
## [1] "multiple meet class req"
## [1] "CRAIG L HURST MD (LAYTON)" "CRAIG L HURST"
## [1] "multiple meet class req"
## [1] "ALEXANDRA GROSVENOR ELLER" "MICHAEL S ESPLIN"
## [1] "multiple meet class req"
## [1] "STEVEN C MEEK" "CAMILLE STEVENSON"
## [1] "multiple meet class req"
## [1] "SCOTT S. REES" "JEFFREY CHARLES BROBERG"
## [1] "multiple meet class req"
## [1] "AMELIA PARRETT" "TROY FLINT PORTER"
## [1] "multiple meet class req"
## [1] "DAVID WARE BRANCH" "AMELIA PARRETT"
## [1] "multiple meet class req"
## [1] "ALTON D BURGETT" "CARA C HEUSER"
## [1] "multiple meet class req"
## [1] "DAVID WARE BRANCH" "MELISSA ANN BROWN"
## [1] "multiple meet class req"
## [1] "CYNTHIA CANNON" "NANCY C ROSE"
## [1] "multiple meet class req"
## [1] "ROBERT O AAGARD" "TENNILLE CLOWARD"
## [1] "multiple meet class req"
## [1] "SCOTT R. JACOB" "DONNA DIZON-TOWNSON"
## [1] "multiple meet class req"
## [1] "KRISTEN HAAS BLACK" "MICHAEL S ESPLIN"
## [1] "multiple meet class req"
## [1] "COBY THOMAS BROWN" "TRACY W WINWARD"
## [1] "multiple meet class req"
## [1] "REED J SKINNER" "HELEN FELTOVICH"
## [1] "multiple meet class req"
## [1] "EVAN JONES" "DONNA DIZON-TOWNSON"
## [1] "multiple meet class req"
## [1] "SEAN HASKETT" "RYAN C. OLLERTON"
## [1] "multiple meet class req"
## [1] "ALEXANDRA GROSVENOR ELLER" "LISA MARIE GRAVELLE"
## [1] "multiple meet class req"
## [1] "SCOTT R. JACOB" "JEFFOREY R. THORPE"
## [1] "multiple meet class req"
## [1] "JEFFREY CHARLES BROBERG" "HELEN FELTOVICH"
## [1] "multiple meet class req"
## [1] "SHANNON L TILLY" "JULIE GAINER"
## [1] "multiple meet class req"
## [1] "LISA MARIE GRAVELLE" "CARA C HEUSER"
## [1] "multiple meet class req"
## [1] "DANIEL FRED KAELBERER" "TROY FLINT PORTER"
## [1] "multiple meet class req"
## [1] "SUSAN MARIE HOWEY" "KATHRYN E WALKER"
## [1] "multiple meet class req"
## [1] "JULIE GAINER" "NATALIE K LOEWEN"
## [1] "multiple meet class req"
## [1] "ALEXANDRA GROSVENOR ELLER" "ROBERT MERRILL"
## [1] "multiple meet class req"
## [1] "DAVID BIERER" "ROBERT L ANDRES"
## [1] "multiple meet class req"
## [1] "AMELIA PARRETT" "LISA MARIE GRAVELLE"
## [1] "multiple meet class req"
## [1] "SCOTT F EPSTEIN" "DENISE JEAN BOUDREAUX-NIPPERT"
## [1] "multiple meet class req"
## [1] "MARGIT SZABO LISTER" "ROBERT L ANDRES"
## [1] "multiple meet class req"
## [1] "ROBERT O AAGARD" "SEAN HASKETT"
## [1] "multiple meet class req"
## [1] "IBRAHIM A HAMMAD" "CARA C HEUSER"
## [1] "multiple meet class req"
## [1] "SPENCER EDMOND PIERSON" "MICHAEL S ESPLIN"
## [1] "multiple meet class req"
## [1] "SCOTT R. JACOB" "JULIE GAINER"
## [1] "multiple meet class req"
## [1] "JENNA MADSEN" "EMMA J MILLER"birth_vag_bq <- birth_vag_bq[,.(
primary_doctor,
primary_doctor_npi,
most_important_fac ,
most_important_fac_npi,
procedure_type,
procedure_modifier,
tp_med_med,
tp_med_surg,
tp_med_medi,
tp_med_path,
tp_med_radi,
tp_med_anes,
tp_med_faci,
ingest_date = Sys.Date()
)]bq_table_upload(x=procedure_table, values= birth_vag_bq, create_disposition='CREATE_IF_NEEDED', write_disposition='WRITE_APPEND')birth_csec
birth_csec %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.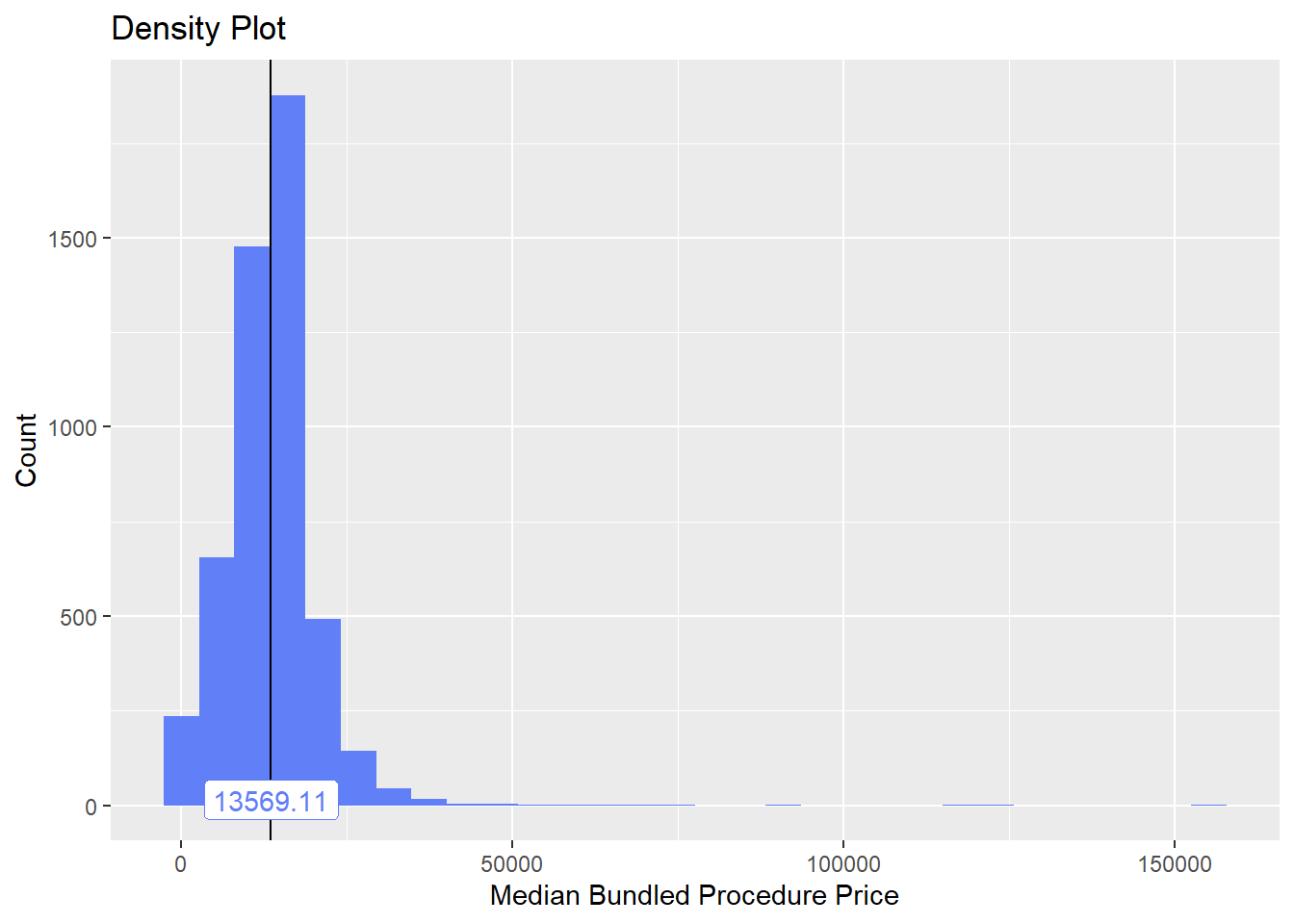
birth_csec %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 287 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.587
## 2 anes_bun_t_anes 0.326
## 3 anes_bun_t_anesth 0.325
## 4 anes_bun_t_deliver 0.324
## 5 anes_bun_t_liver 0.324
## 6 anes_bun_t_cs 0.291
## 7 faci_bun_t_initial 0.235
## 8 anes_bun_t_vag 0.188
## 9 anes_bun_t_analg 0.183
## 10 anes_bun_t_add - on 0.174
## # ... with 277 more rowsbirth_csec <- birth_csec[,`:=`(
surg_sp_name_clean = surg_sp_npi %>% map_chr(get_npi_standard_name),
surg_bp_name_clean = surg_bp_npi %>% map_chr(get_npi_standard_name),
medi_sp_name_clean = medi_sp_npi %>% map_chr(get_npi_standard_name),
medi_bp_name_clean = medi_bp_npi %>% map_chr(get_npi_standard_name),
radi_sp_name_clean = radi_sp_npi %>% map_chr(get_npi_standard_name),
radi_bp_name_clean = radi_bp_npi %>% map_chr(get_npi_standard_name),
path_sp_name_clean = path_sp_npi %>% map_chr(get_npi_standard_name),
path_bp_name_clean = path_bp_npi %>% map_chr(get_npi_standard_name),
anes_sp_name_clean = anes_sp_npi %>% map_chr(get_npi_standard_name),
anes_bp_name_clean = anes_bp_npi %>% map_chr(get_npi_standard_name),
faci_sp_name_clean = faci_sp_npi %>% map_chr(get_npi_standard_name),
faci_bp_name_clean = faci_bp_npi %>% map_chr(get_npi_standard_name)
)]birth_csec %>% saveRDS("birth_csec.RDS")birth_csec <- readRDS("birth_csec.RDS")birth_csec[cnt>5 & faci_bun_sum_med > 3000 & tp_med < 60000] %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.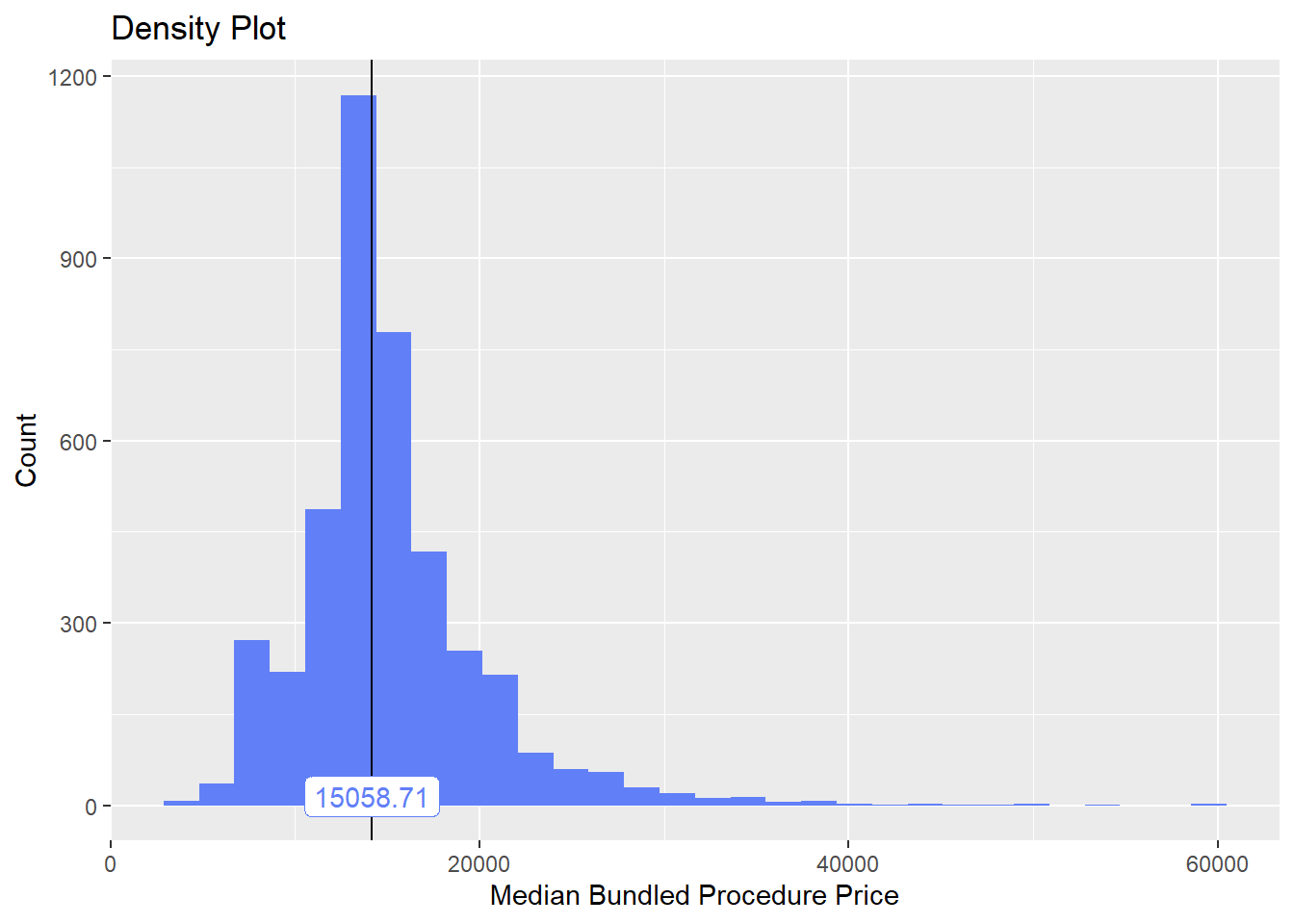
birth_csec[cnt>5 & faci_bun_sum_med > 3000 & tp_med < 60000] %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 272 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.299
## 2 faci_bun_t_initial 0.27
## 3 faci_bun_t_visit 0.186
## 4 anes_bun_t_vag 0.176
## 5 anes_bun_t_analg 0.174
## 6 faci_bun_t_dept 0.161
## 7 faci_bun_t_emergency 0.161
## 8 anes_bun_t_add - on 0.158
## 9 radi_bun_t_chest 0.147
## 10 faci_bun_t_discharge 0.132
## # ... with 262 more rowsbirth_csec <- birth_csec[cnt>5 & faci_bun_sum_med > 3000 & tp_med < 60000]birth_csec %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.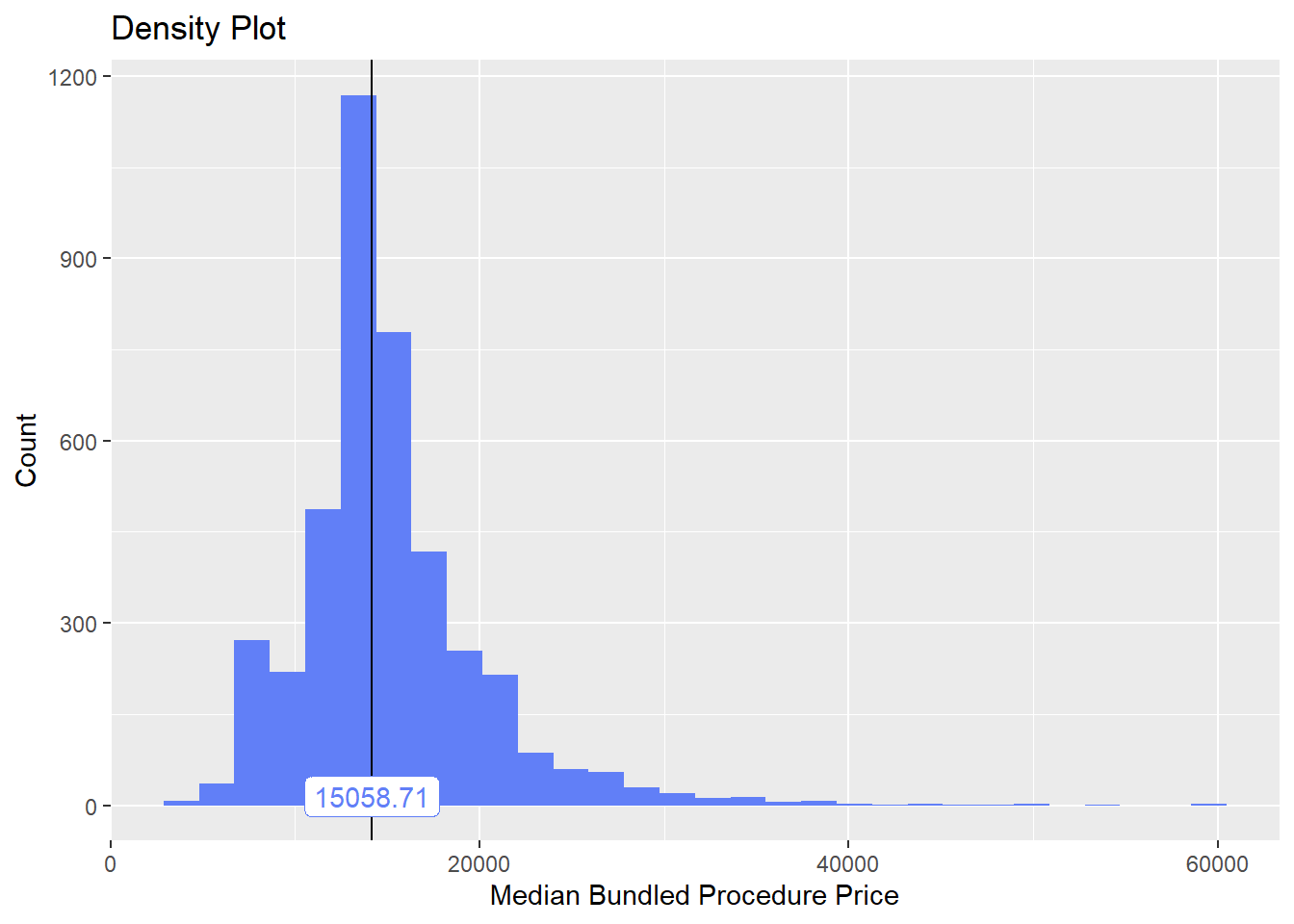
birth_csec %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 272 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.299
## 2 faci_bun_t_initial 0.27
## 3 faci_bun_t_visit 0.186
## 4 anes_bun_t_vag 0.176
## 5 anes_bun_t_analg 0.174
## 6 faci_bun_t_dept 0.161
## 7 faci_bun_t_emergency 0.161
## 8 anes_bun_t_add - on 0.158
## 9 radi_bun_t_chest 0.147
## 10 faci_bun_t_discharge 0.132
## # ... with 262 more rowsbirth_csec %>% get_tag_density_information("faci_bun_t_initial") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ faci_bun_t_initial"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 1440## $dist_plots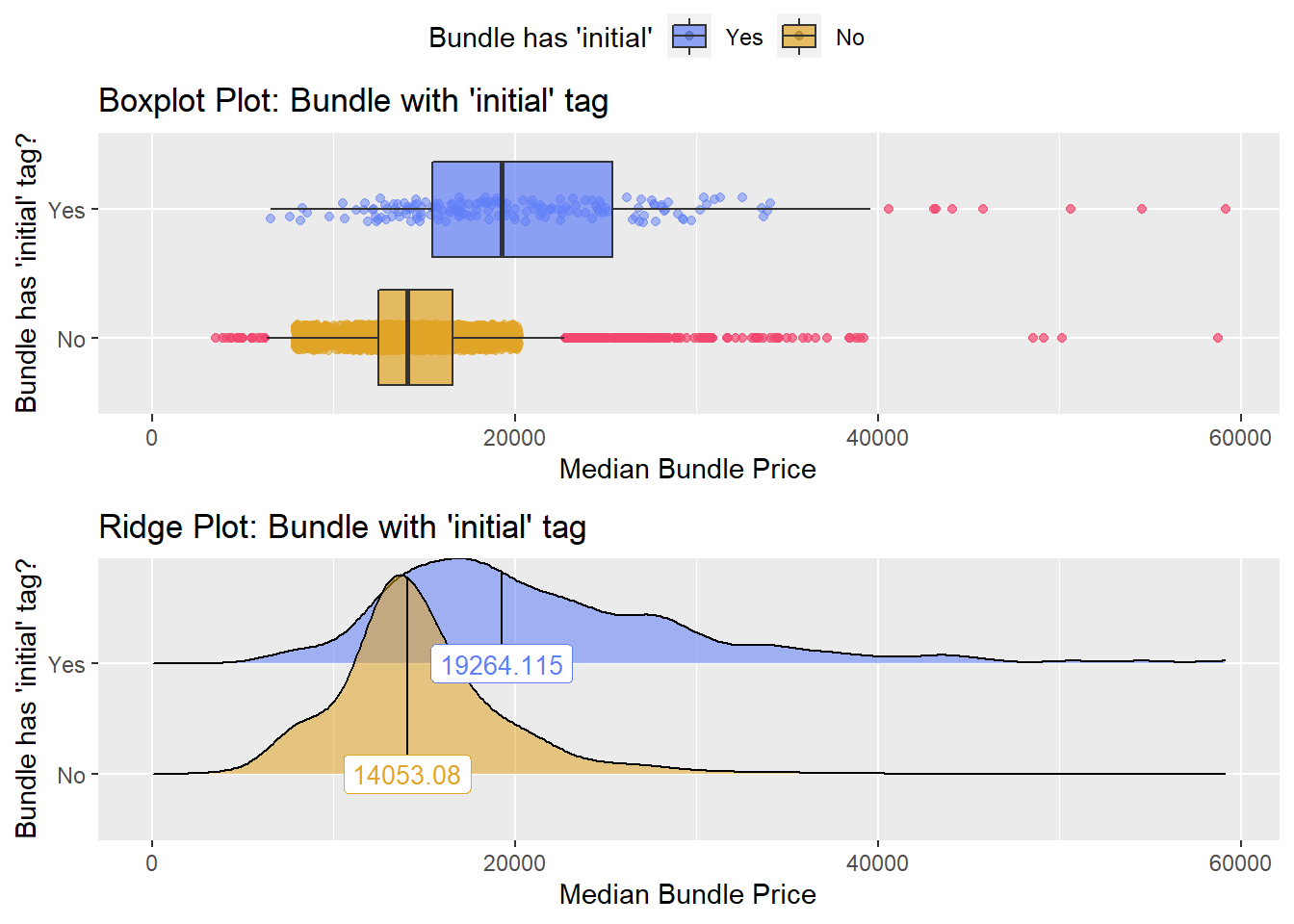
##
## $stat_tables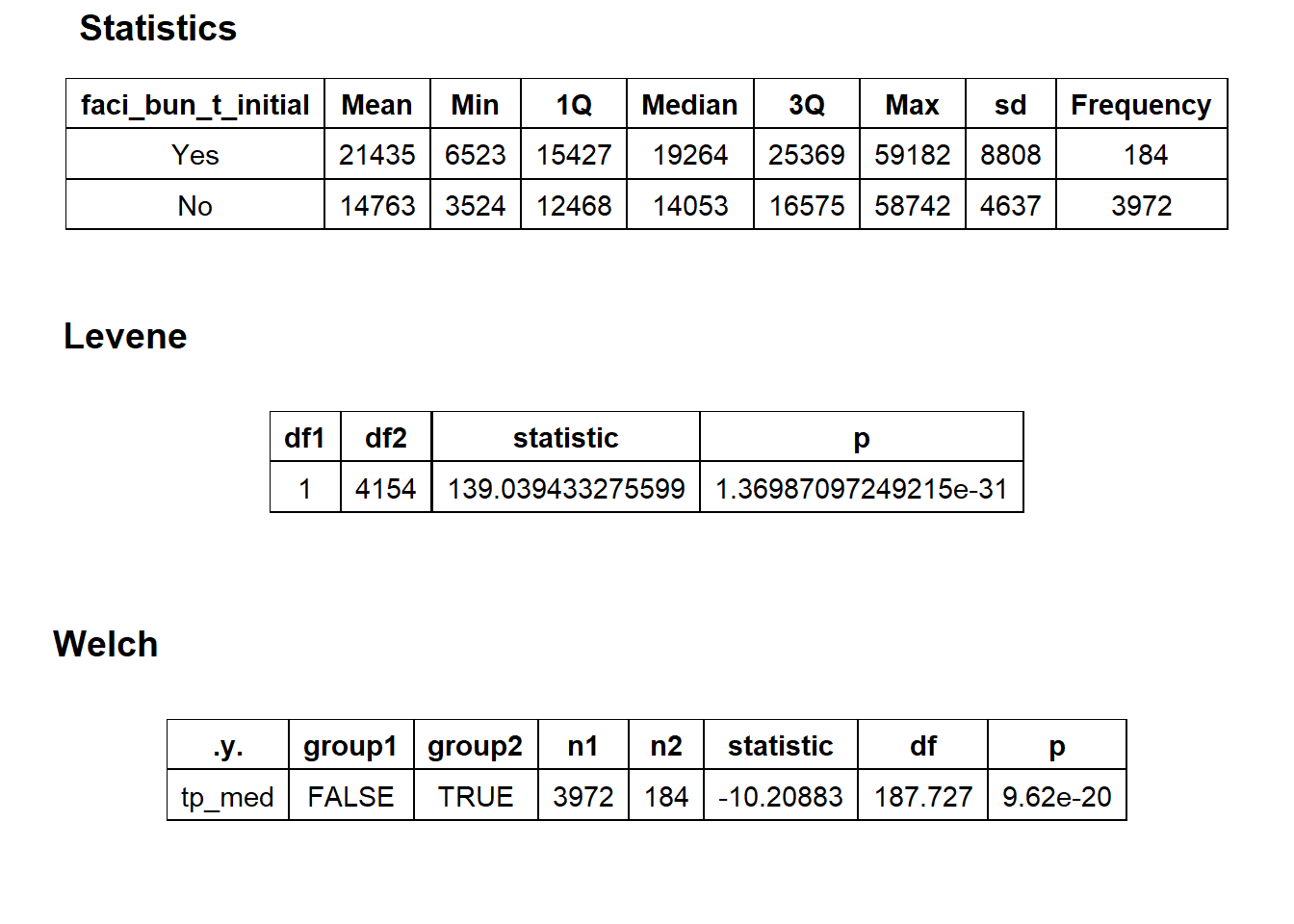
Statiscally significant different groups, but too small to represent own group.
birth_csec <- birth_csec[faci_bun_t_initial==F]Now, we wil recheck the median price density and highly correlated tags.
birth_csec %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.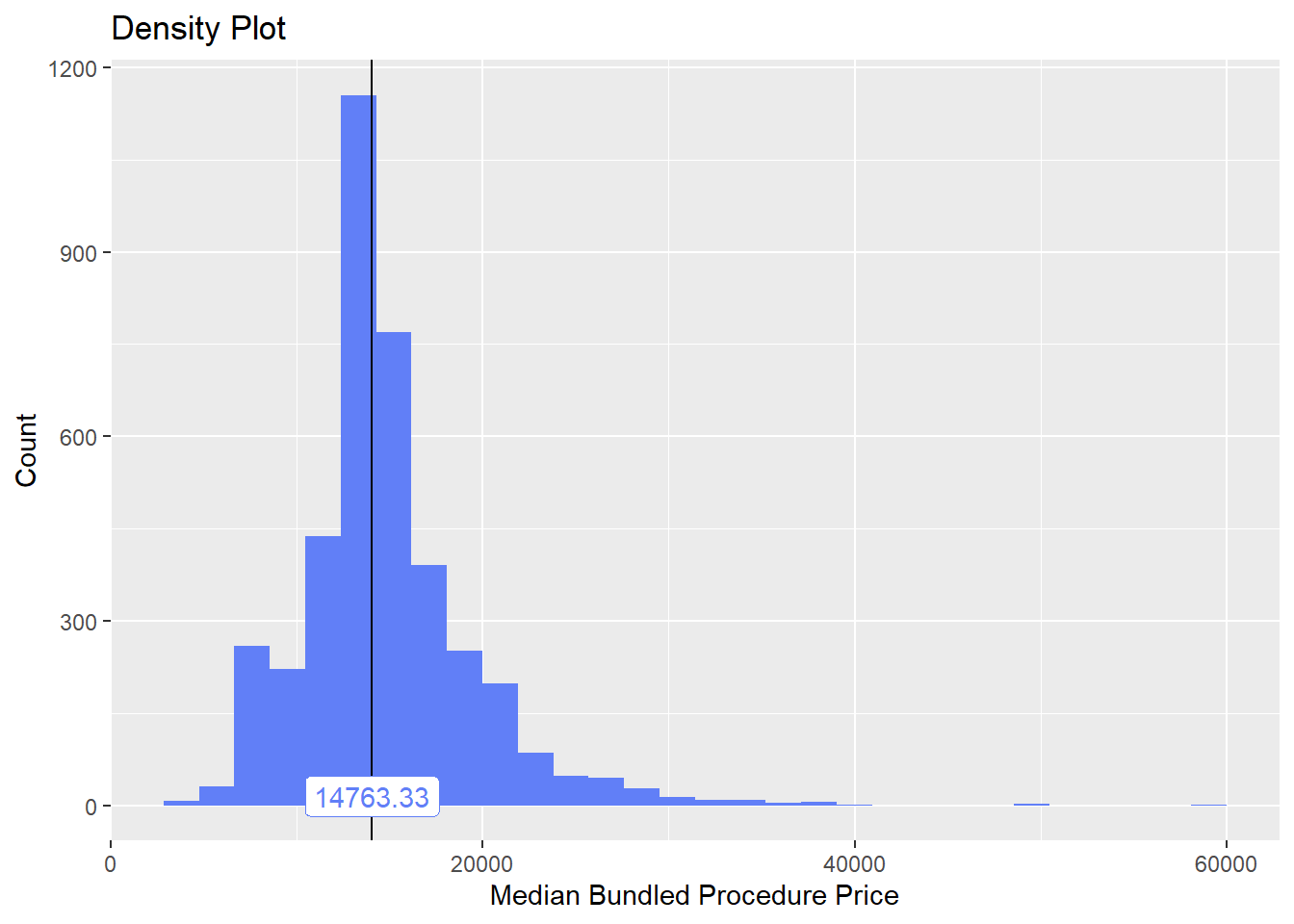
birth_csec %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 231 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.197
## 2 anes_bun_t_vag 0.180
## 3 anes_bun_t_analg 0.178
## 4 anes_bun_t_add - on 0.161
## 5 faci_bun_t_visit 0.147
## 6 faci_bun_t_dept 0.128
## 7 faci_bun_t_emergency 0.128
## 8 path_bun_t_complete 0.0935
## 9 faci_bun_t_discharge 0.0921
## 10 surg_bun_t_remove 0.0916
## # ... with 221 more rowsbirth_csec %>% get_tag_density_information("faci_bun_t_emergency") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ faci_bun_t_emergency"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 804## $dist_plots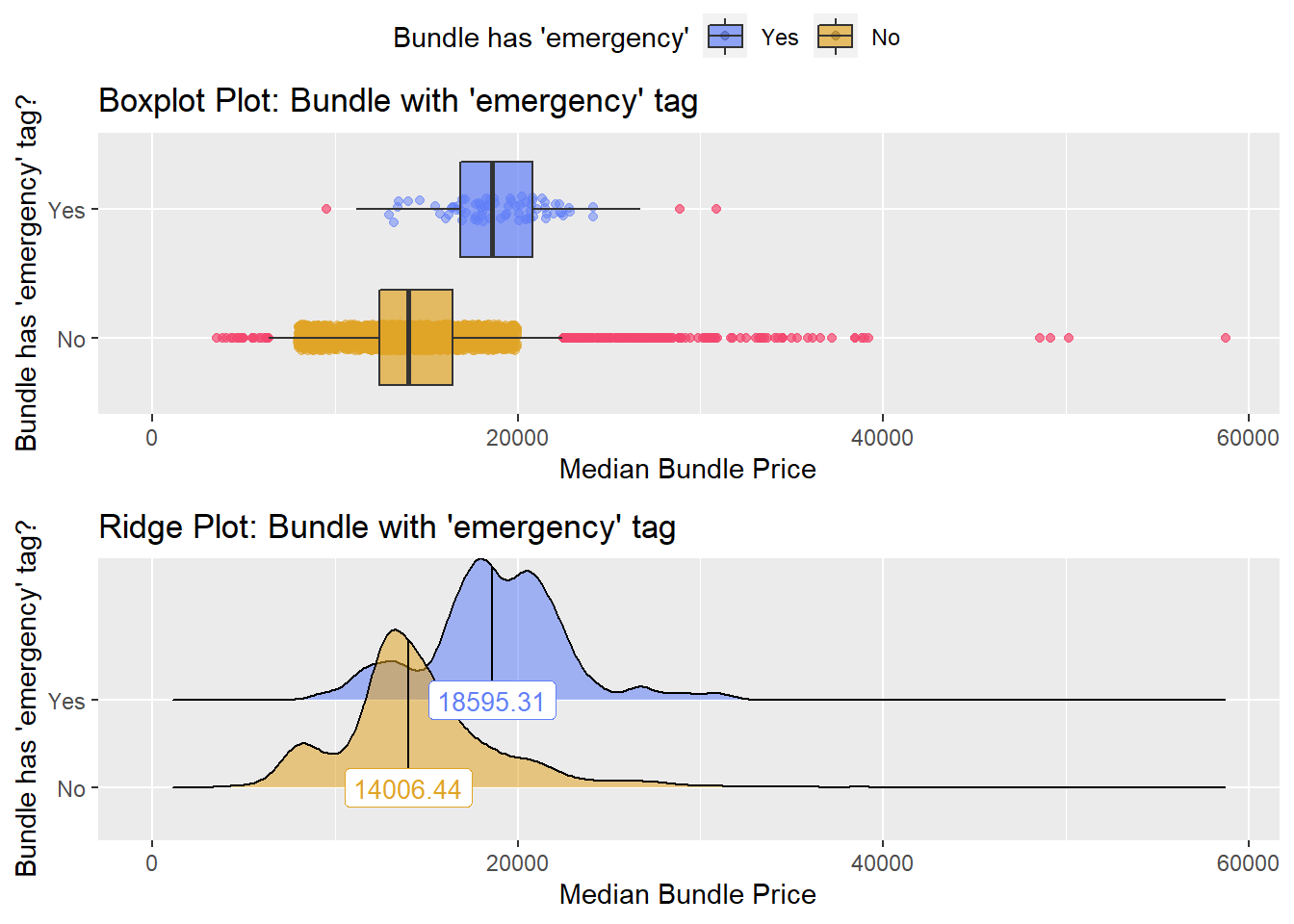
##
## $stat_tables
birth_csec <- birth_csec[faci_bun_t_emergency == F & faci_bun_t_discharge == F]ow, we wil recheck the median price density and highly correlated tags.
birth_csec %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
birth_csec %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 224 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.192
## 2 anes_bun_t_vag 0.177
## 3 anes_bun_t_analg 0.175
## 4 anes_bun_t_add - on 0.159
## 5 surg_bun_t_remove 0.0961
## 6 faci_bun_t_hospital 0.0908
## 7 faci_bun_t_subsequent 0.0881
## 8 faci_bun_t_care 0.0866
## 9 path_bun_t_complete 0.0847
## 10 faci_bun_t_observation 0.0844
## # ... with 214 more rowsbirth_csec %>% get_tag_density_information("anes_bun_t_vag") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ anes_bun_t_vag"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 589## $dist_plots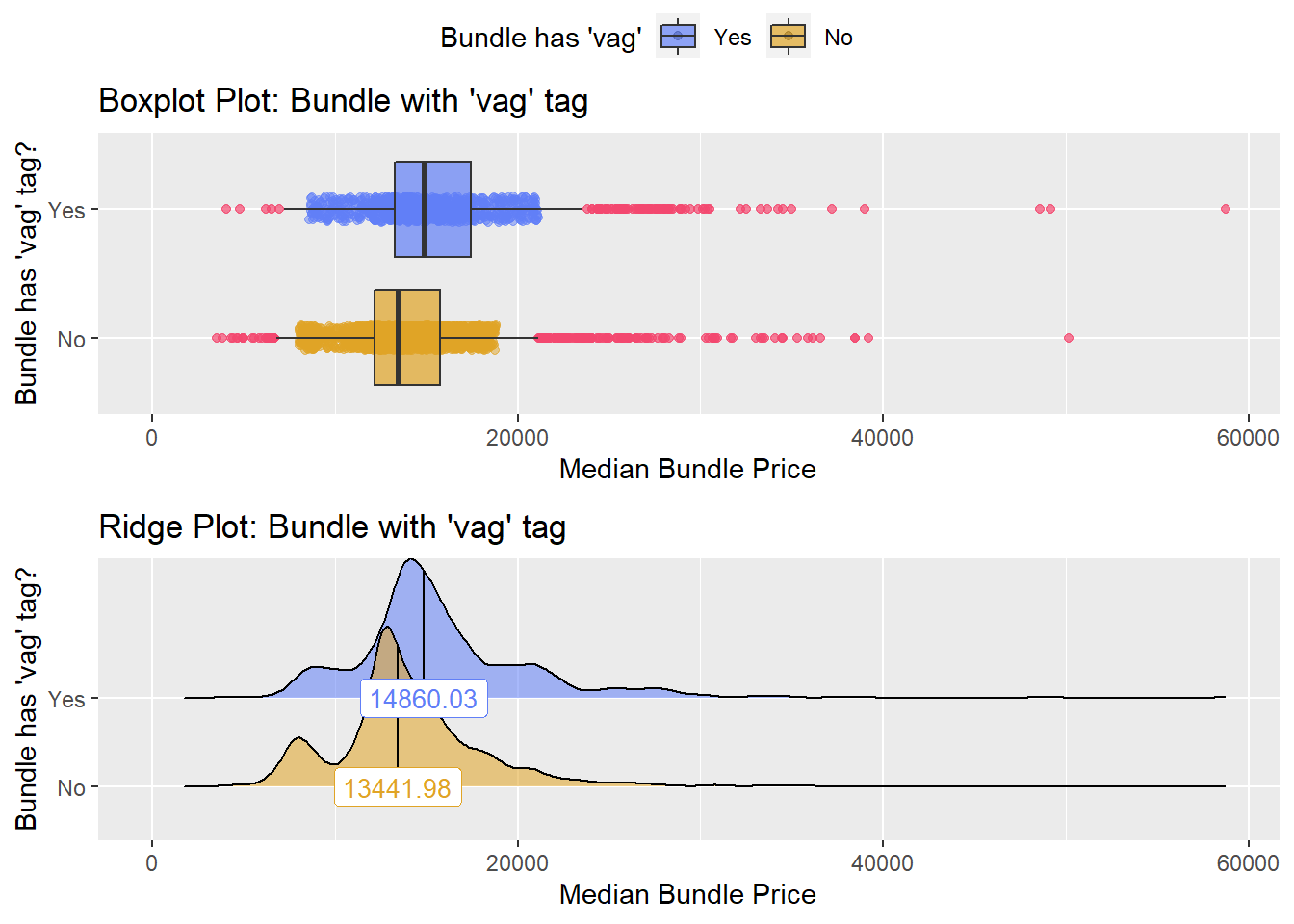
##
## $stat_tables
birth_csec_w_anes <- birth_csec[anes_bun_t_vag==T]
birth_csec_n_anes <- birth_csec[anes_bun_t_vag==F]birth_csec_n_anes
birth_csec_n_anes %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.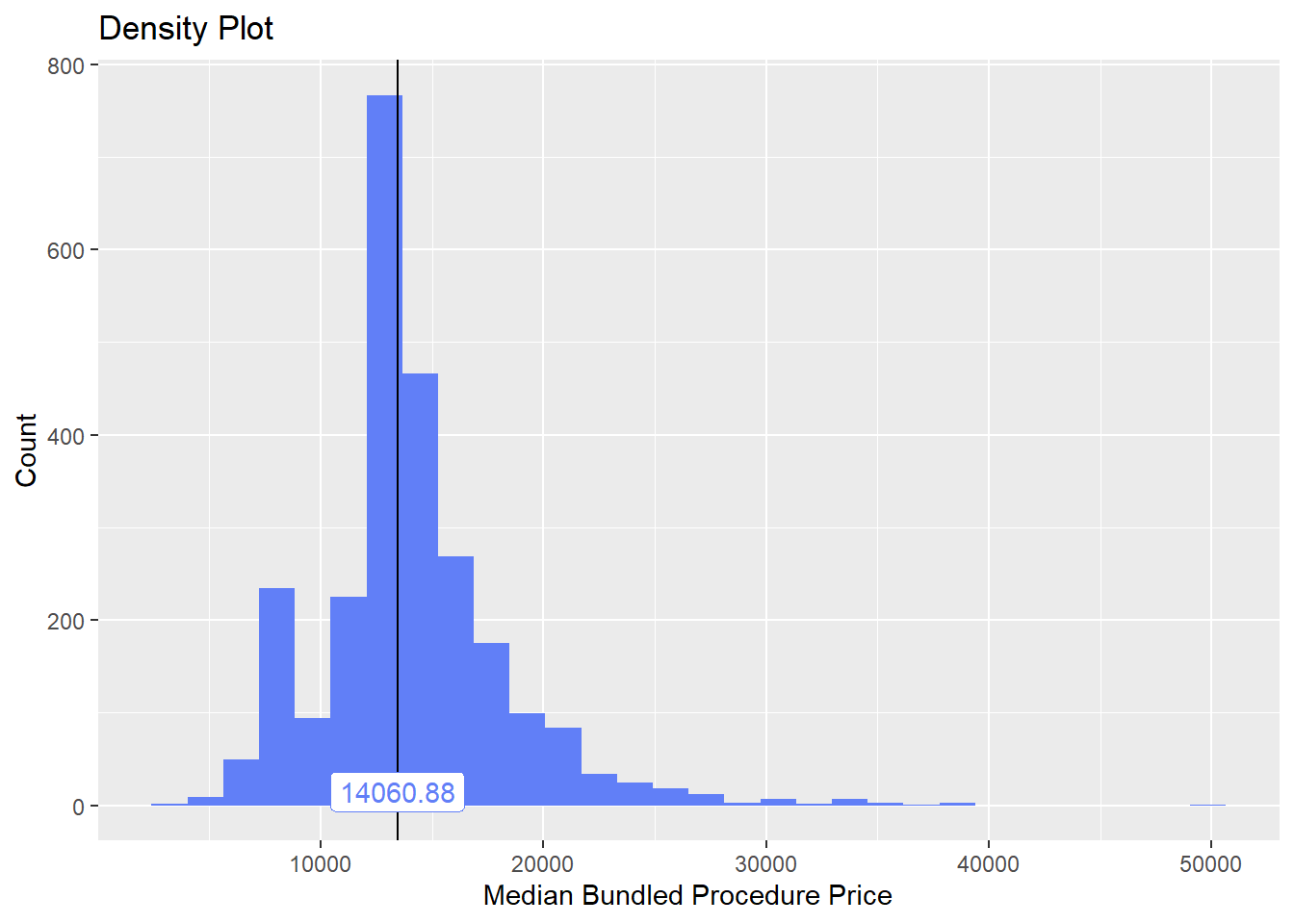
birth_csec_n_anes %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 204 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.188
## 2 medi_bun_t_puncture 0.128
## 3 medi_bun_t_routine 0.128
## 4 medi_bun_t_venipuncture 0.128
## 5 faci_bun_t_observation 0.112
## 6 path_bun_t_antibod 0.102
## 7 surg_bun_t_bladder 0.1
## 8 path_bun_t_screen 0.0978
## 9 path_bun_t_complete 0.096
## 10 path_bun_t_anti 0.0958
## # ... with 194 more rowsbirth_csec_w_anes
we will now look at the density distribution for csection births with anesthesia
birth_csec_w_anes %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.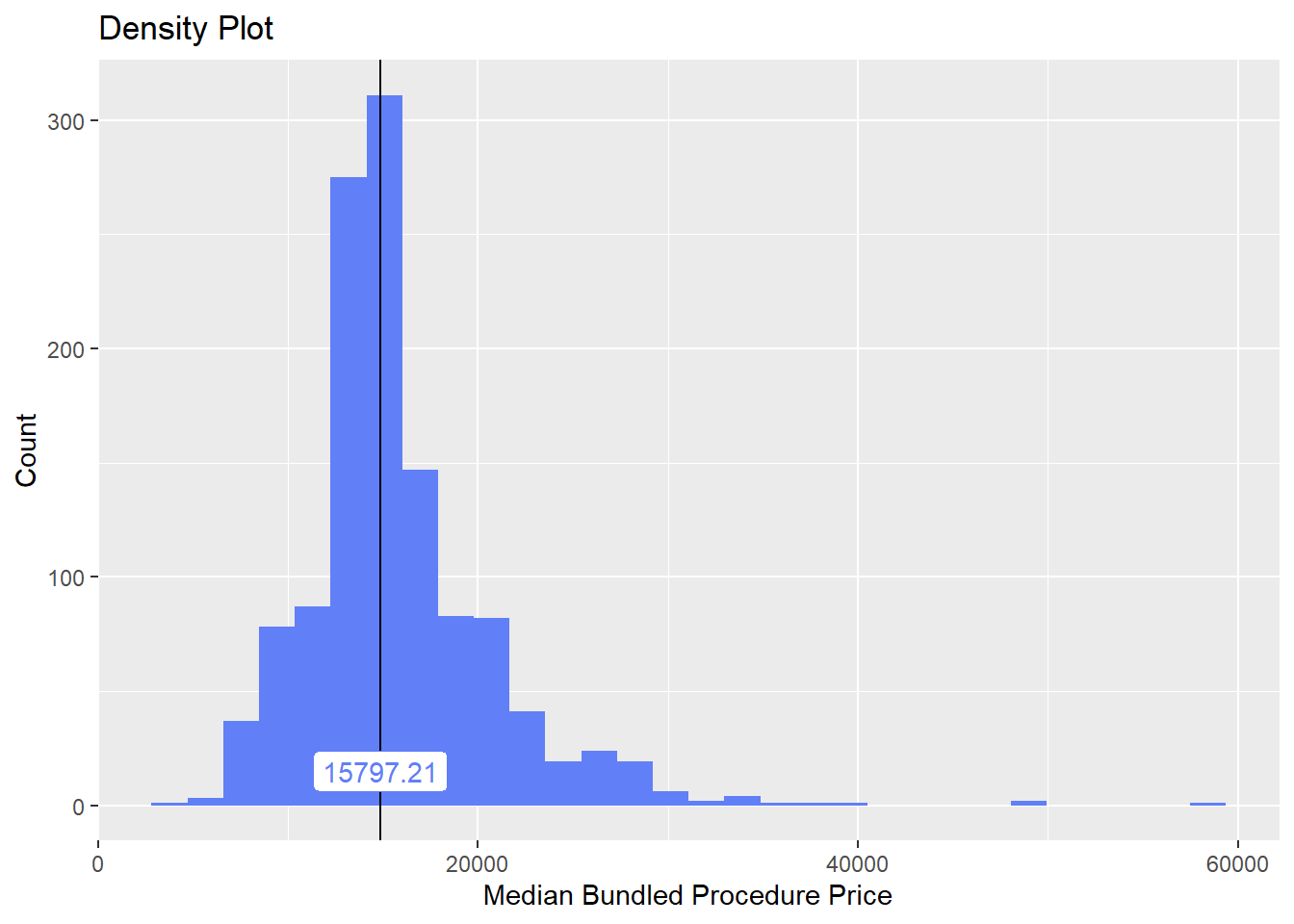
birth_csec_w_anes %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 152 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_remove 0.248
## 2 surg_bun_t_cyst 0.161
## 3 surg_bun_t_ovarian 0.161
## 4 surg_bun_t_remov 0.161
## 5 surg_bun_t_removal 0.161
## 6 anes_bun_t_analg 0.157
## 7 duration_max 0.127
## 8 radi_bun_t_artery 0.116
## 9 faci_bun_t_hospital 0.105
## 10 faci_bun_t_subsequent 0.105
## # ... with 142 more rowsbirth_csec_w_anes[surg_bun_t_remove==T] %>% nrow()## [1] 1only one in with that tag
birth_csec_w_anes <- birth_csec_w_anes[surg_bun_t_remove==F]birth_csec_w_anes[surg_bun_t_ovarian==T] %>% nrow()## [1] 1only one in with that tag
birth_csec_w_anes <- birth_csec_w_anes[surg_bun_t_ovarian==F]we will now look at the density distribution for csection births with anesthesia
birth_csec_w_anes %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.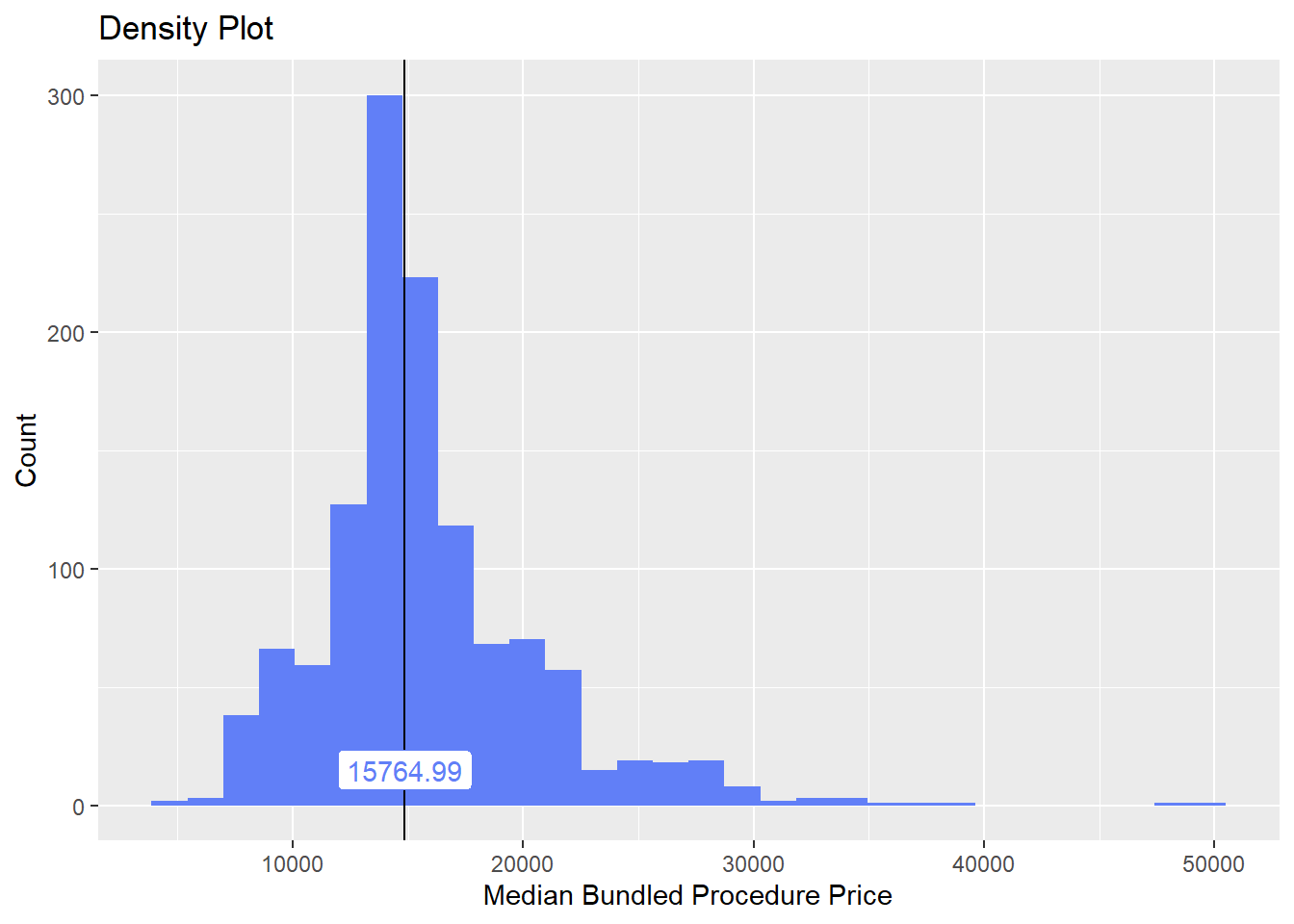
birth_csec_w_anes %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 144 x 2
## name correlation
## <chr> <dbl>
## 1 radi_bun_t_artery 0.120
## 2 faci_bun_t_hospital 0.117
## 3 faci_bun_t_subsequent 0.117
## 4 faci_bun_t_care 0.114
## 5 radi_bun_t_echo 0.0951
## 6 path_bun_t_patho 0.0927
## 7 path_bun_t_pathologist 0.0927
## 8 path_bun_t_tissue 0.0927
## 9 path_bun_t_virus 0.0879
## 10 path_bun_t_complete 0.0877
## # ... with 134 more rowsbirth_csec %>% get_tag_density_information("radi_bun_t_artery") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ radi_bun_t_artery"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 1320## $dist_plots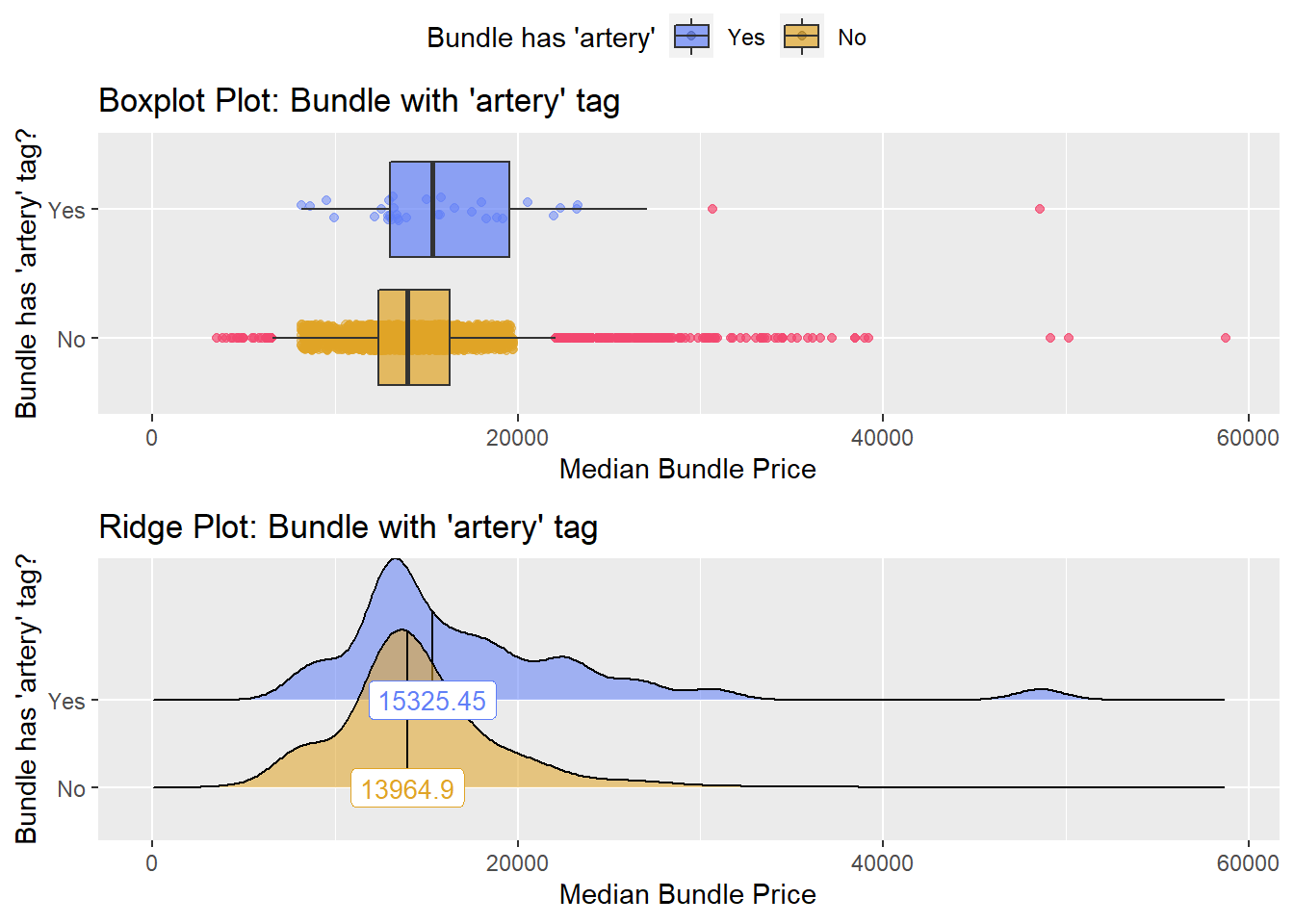
##
## $stat_tables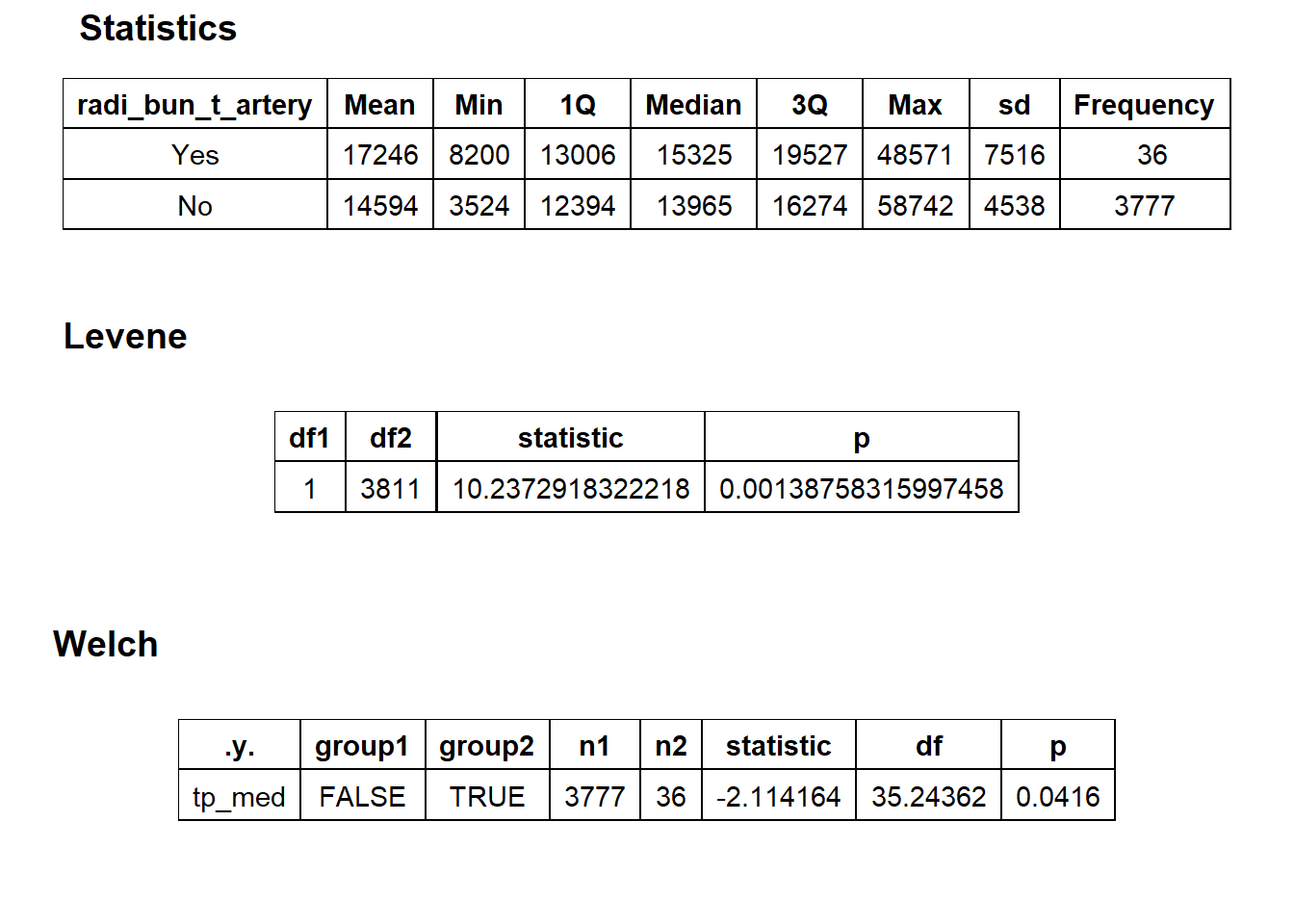
birth_csec_w_anes <- birth_csec_w_anes[radi_bun_t_artery==F]we will now look at the density distribution for csection births with anesthesia
birth_csec_w_anes %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.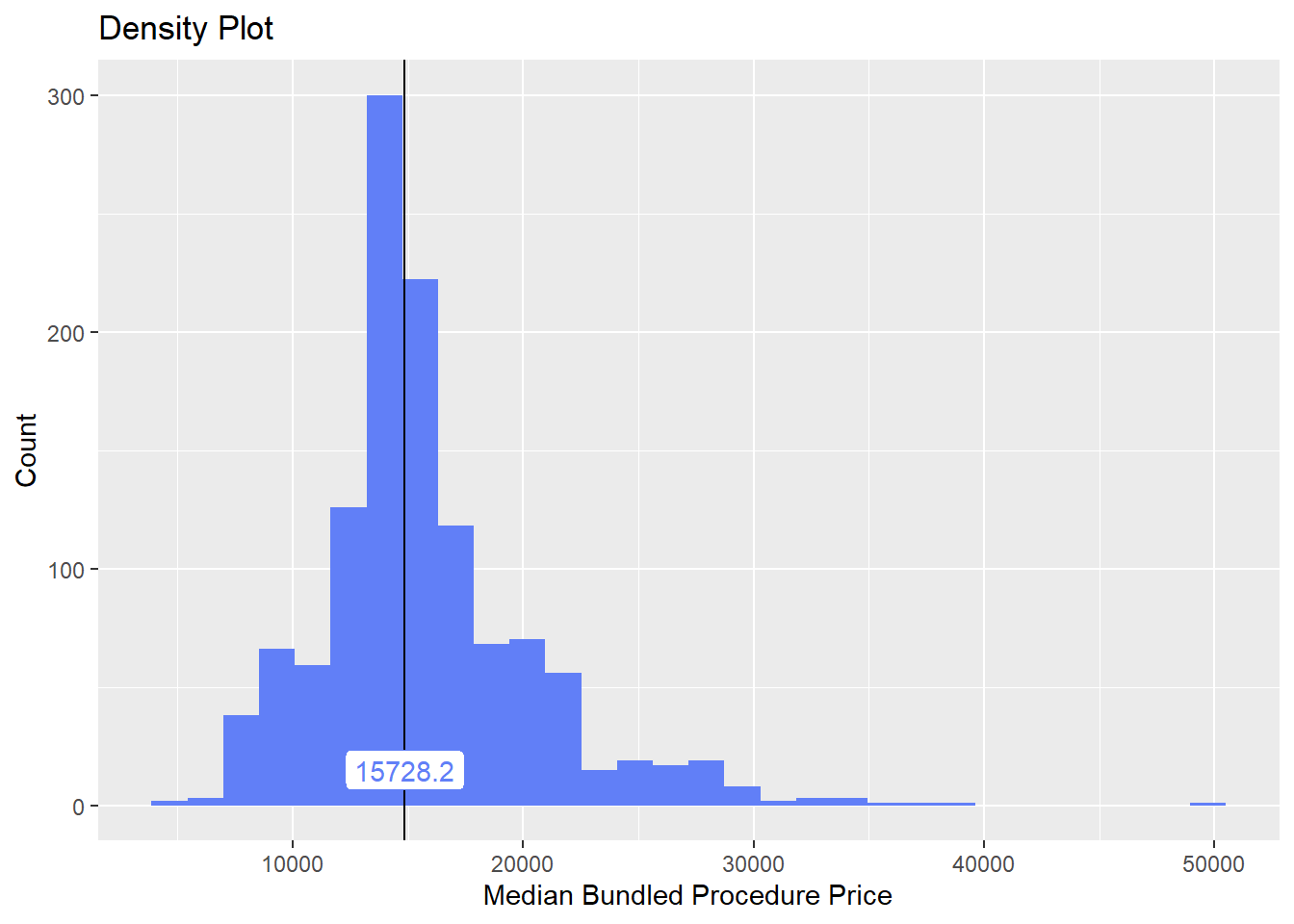
birth_csec_w_anes %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 143 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_hospital 0.125
## 2 faci_bun_t_subsequent 0.125
## 3 faci_bun_t_care 0.123
## 4 path_bun_t_patho 0.0939
## 5 path_bun_t_pathologist 0.0939
## 6 path_bun_t_tissue 0.0939
## 7 path_bun_t_virus 0.0902
## 8 duration_max 0.0897
## 9 path_bun_t_complete 0.0817
## 10 surg_bun_t_abdom 0.0779
## # ... with 133 more rowsbirth_csec_w_anes %>% get_tag_density_information("faci_bun_t_hospital") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ faci_bun_t_hospital"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 827## $dist_plots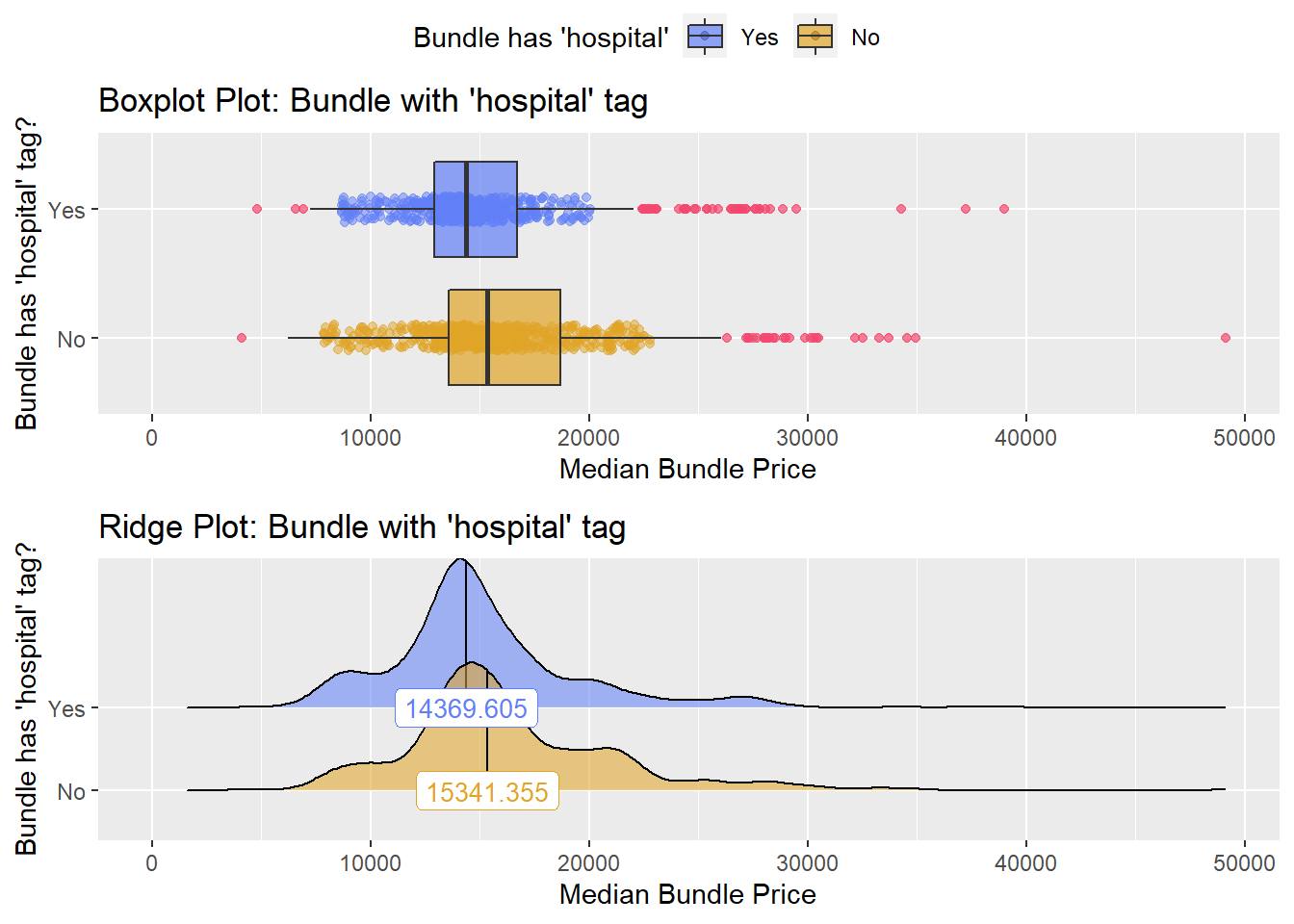
##
## $stat_tables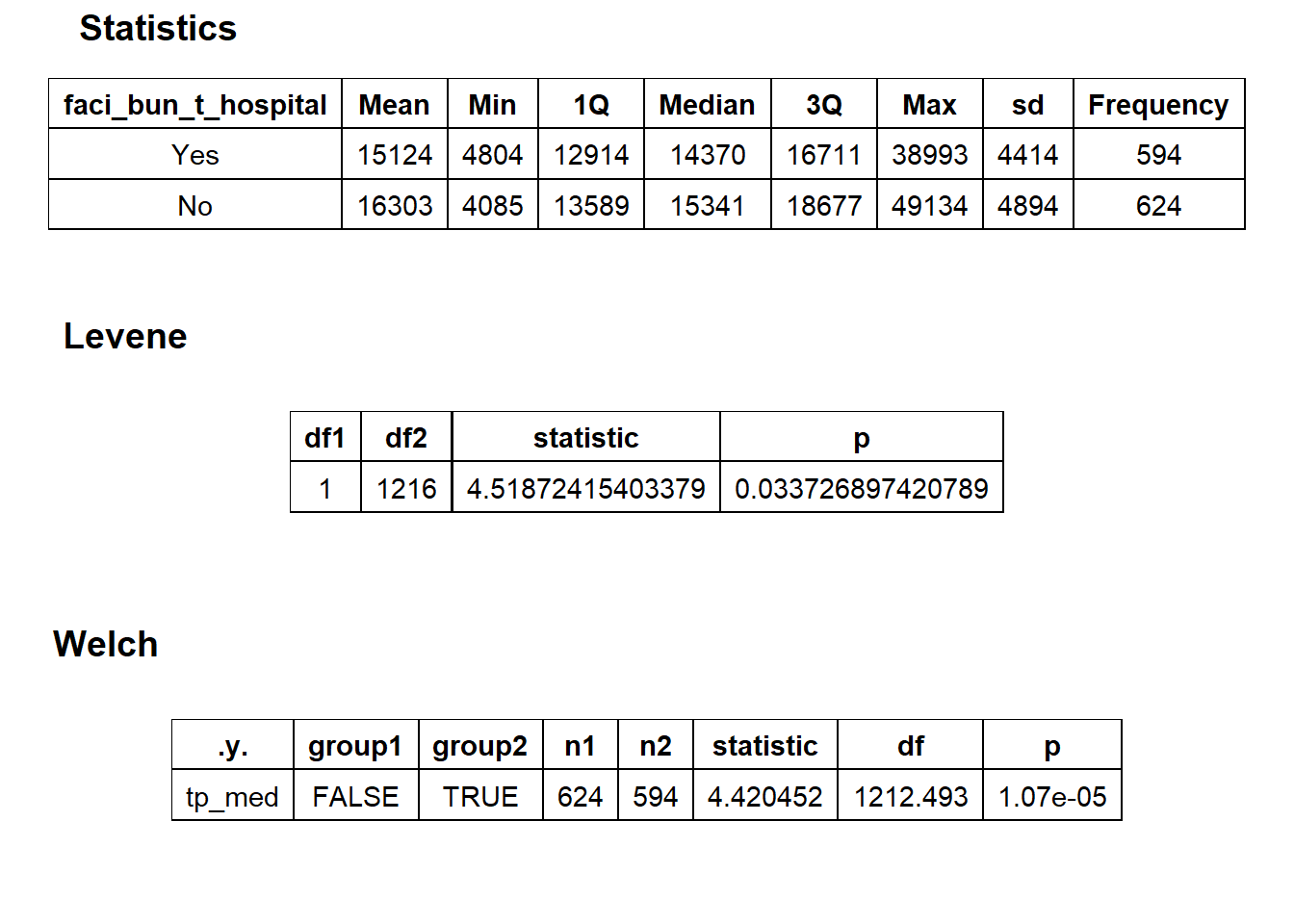
birth_csec_w_anes_w_sub_hosp_care <- birth_csec_w_anes[faci_bun_t_care==T & faci_bun_t_subsequent ==T]
birth_csec_w_anes <- birth_csec_w_anes[faci_bun_t_care==F & faci_bun_t_subsequent ==F]we will now look at the density distribution for csection births with anesthesia
birth_csec_w_anes %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
birth_csec_w_anes %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 109 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.123
## 2 path_bun_t_complete 0.121
## 3 path_bun_t_colon 0.117
## 4 path_bun_t_count 0.117
## 5 path_bun_t_virus 0.117
## 6 surg_bun_t_abd 0.100
## 7 surg_bun_t_abdom 0.100
## 8 surg_bun_t_abdomen 0.100
## 9 surg_bun_t_open 0.100
## 10 surg_bun_t_opening 0.100
## # ... with 99 more rowsbirth_csec_w_anes %>% get_tag_density_information("path_bun_t_complete") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_complete"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 1580## $dist_plots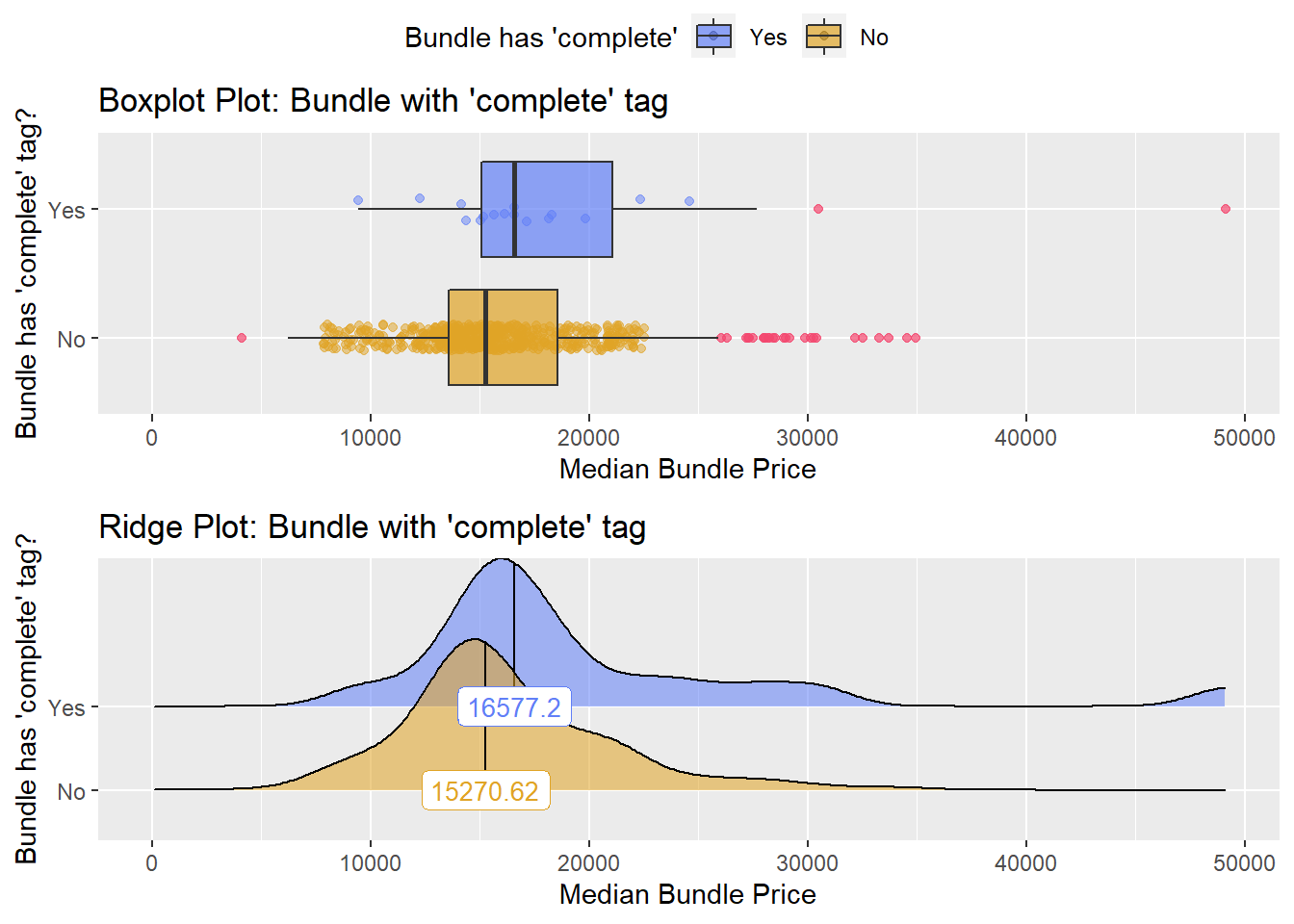
##
## $stat_tables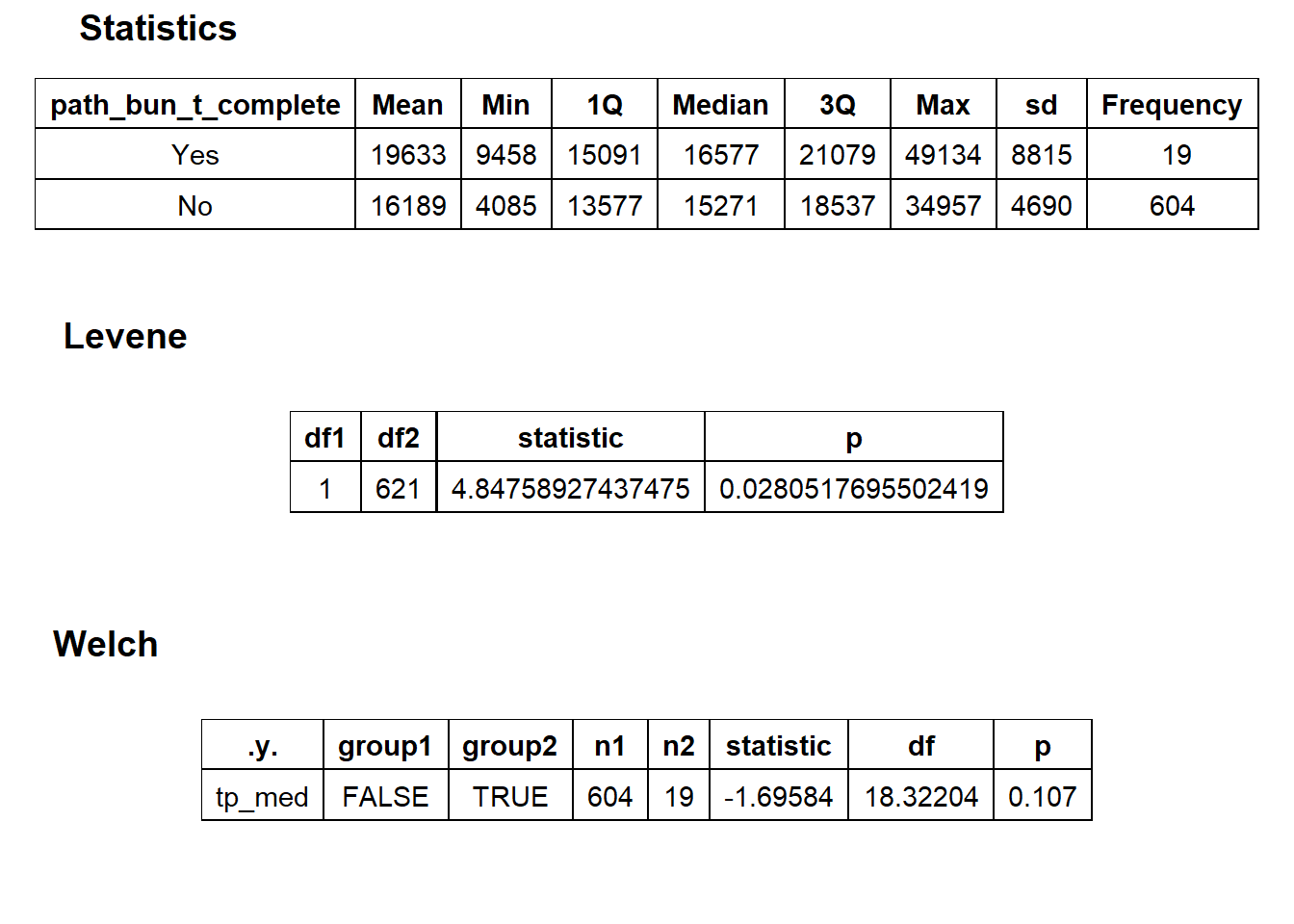
birth_csec_w_anes[path_bun_t_colon==T] %>% nrow()## [1] 1birth_csec_w_anes[surg_bun_t_abdom==T] %>% nrow()## [1] 1There is weak evidence to split, but since only 19 of them are listed a complete pathology panel, I will omit the 19. This also gets rid of a few outliers.
birth_csec_w_anes <- birth_csec_w_anes[path_bun_t_complete==F & path_bun_t_colon==F & surg_bun_t_abdom==F]we will now look at the density distribution for csection births with anesthesia
birth_csec_w_anes %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.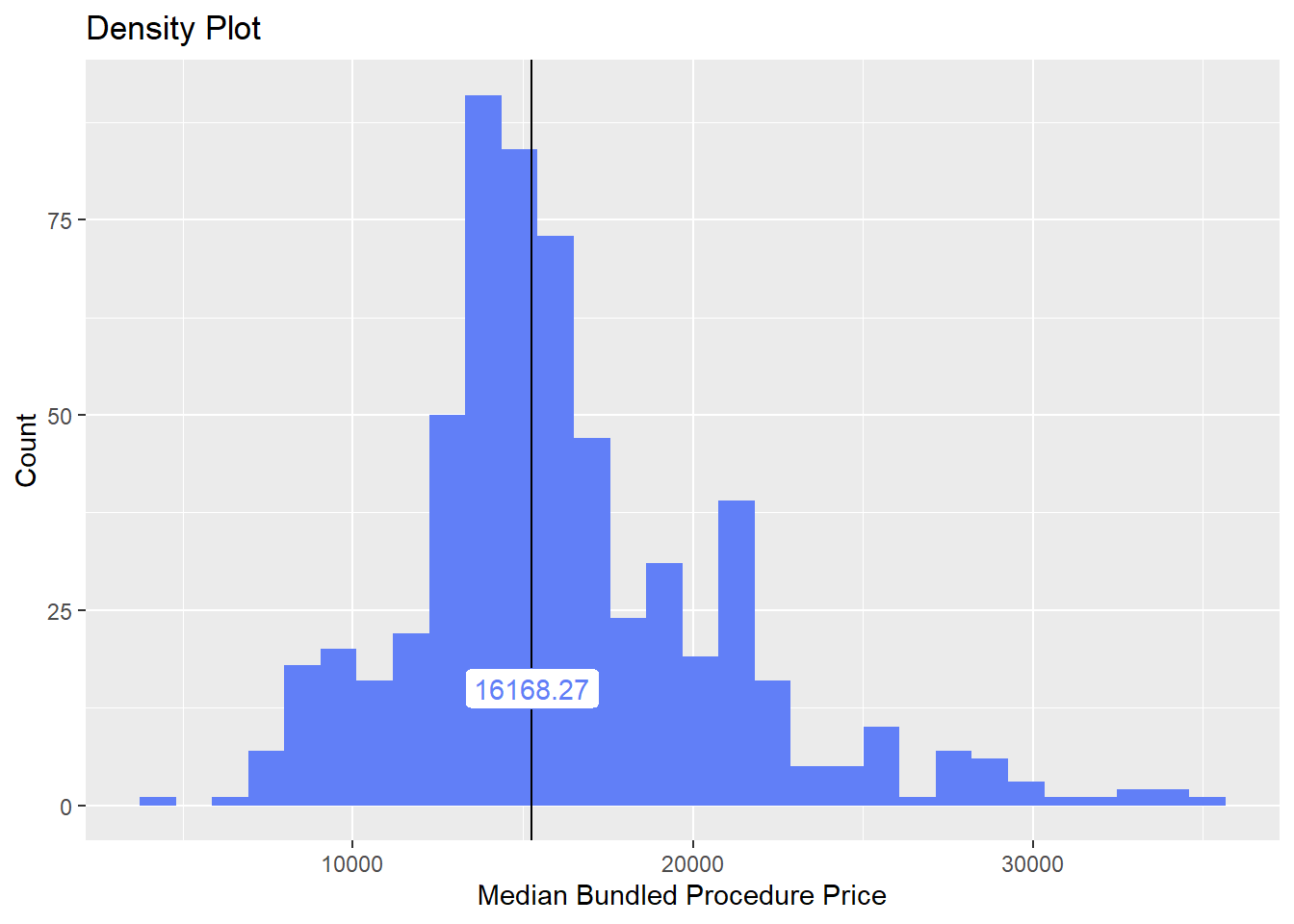
birth_csec_w_anes %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 86 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.108
## 2 surg_bun_t_b9 0.104
## 3 surg_bun_t_exc 0.104
## 4 surg_bun_t_block 0.0932
## 5 surg_bun_t_injection 0.0932
## 6 path_bun_t_amniotic 0.0808
## 7 path_bun_t_protein 0.0808
## 8 path_bun_t_patho 0.0734
## 9 path_bun_t_pathologist 0.0734
## 10 path_bun_t_tissue 0.0734
## # ... with 76 more rowsbirth_csec_w_anes %>% get_tag_density_information("surg_bun_t_exc") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_exc"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 3130## $dist_plots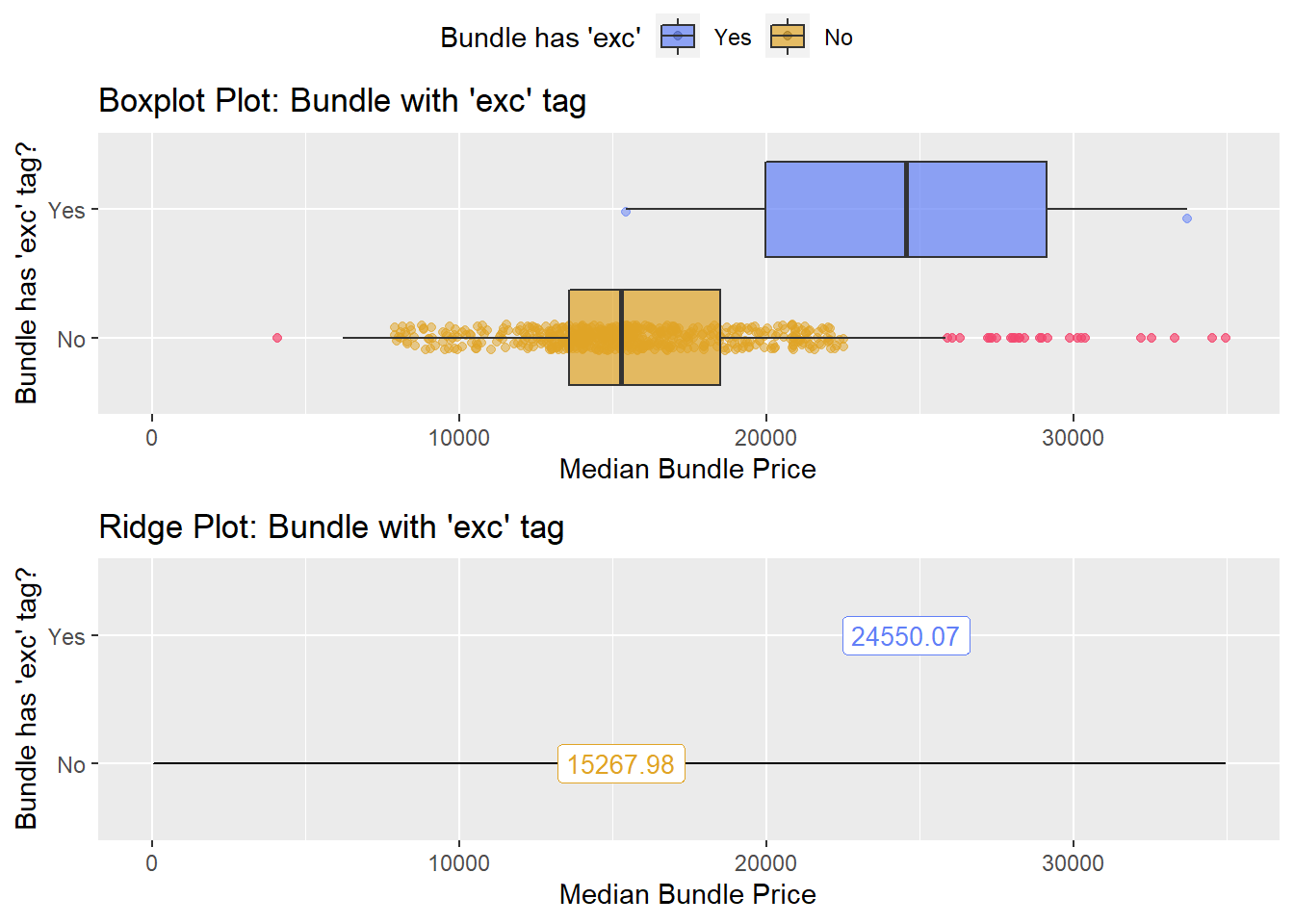
##
## $stat_tables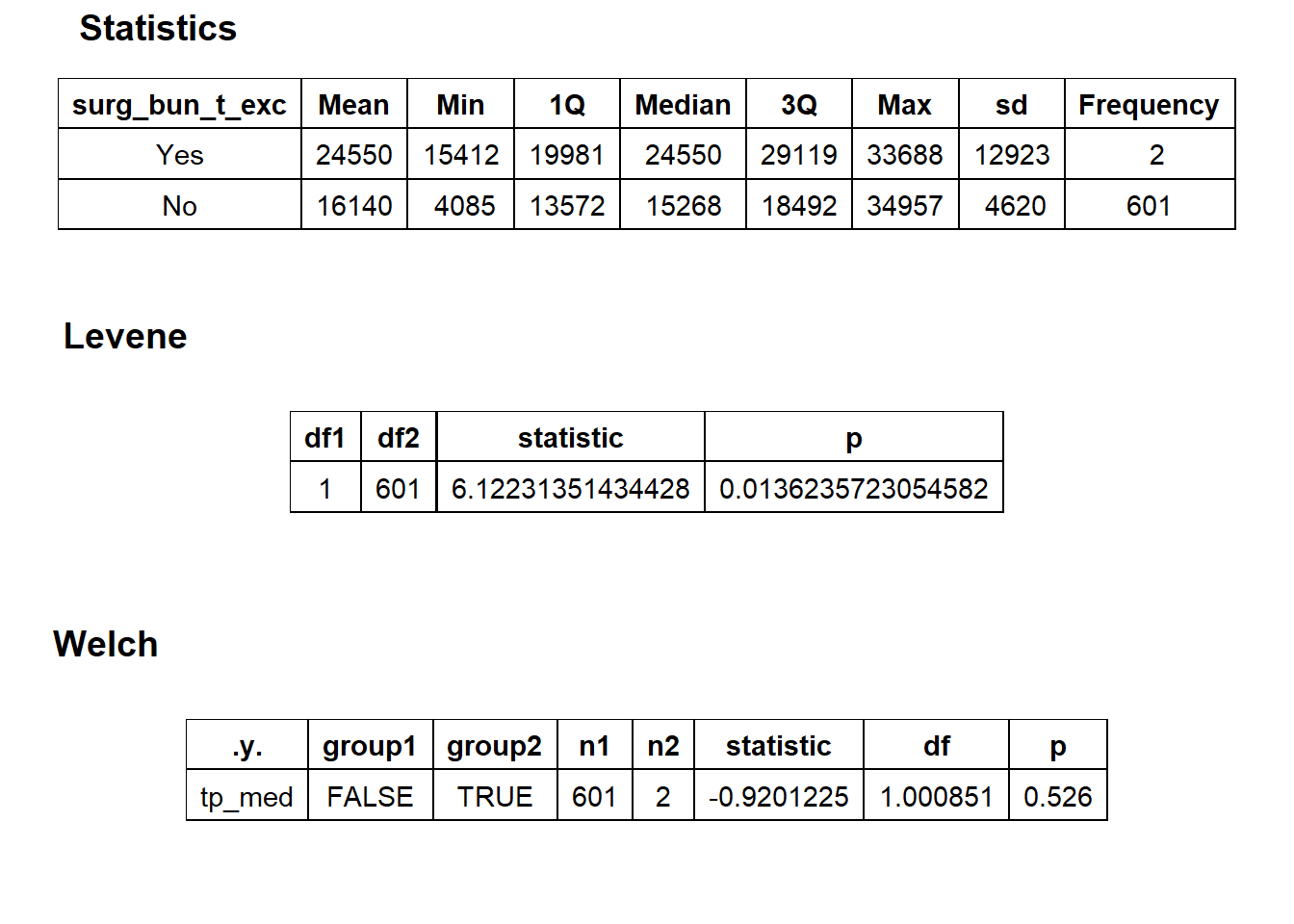 Small subset and not a standard procedure based on frequency.
Small subset and not a standard procedure based on frequency.
birth_csec_w_anes <- birth_csec_w_anes[surg_bun_t_exc==F]we will now look at the density distribution for csection births with anesthesia
birth_csec_w_anes %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
birth_csec_w_anes %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 84 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.106
## 2 surg_bun_t_block 0.0951
## 3 surg_bun_t_injection 0.0951
## 4 path_bun_t_amniotic 0.082
## 5 path_bun_t_protein 0.082
## 6 surg_bun_t_ant 0.065
## 7 surg_bun_t_antepartum 0.065
## 8 surg_bun_t_art 0.065
## 9 surg_bun_t_part 0.065
## 10 surg_bun_t_tum 0.065
## # ... with 74 more rowsbirth_csec_w_anes[surg_bun_t_injection==T] %>% nrow()## [1] 11birth_csec_w_anes[path_bun_t_amniotic==T] %>% nrow()## [1] 1birth_csec_w_anes[surg_bun_t_antepartum==T] %>% nrow()## [1] 40birth_csec_w_anes[surg_bun_t_perineum==T] %>% nrow()## [1] 1birth_csec_w_anes <- birth_csec_w_anes[surg_bun_t_injection==F & path_bun_t_amniotic==F & surg_bun_t_perineum==F]we will now look at the density distribution for csection births with anesthesia
birth_csec_w_anes %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.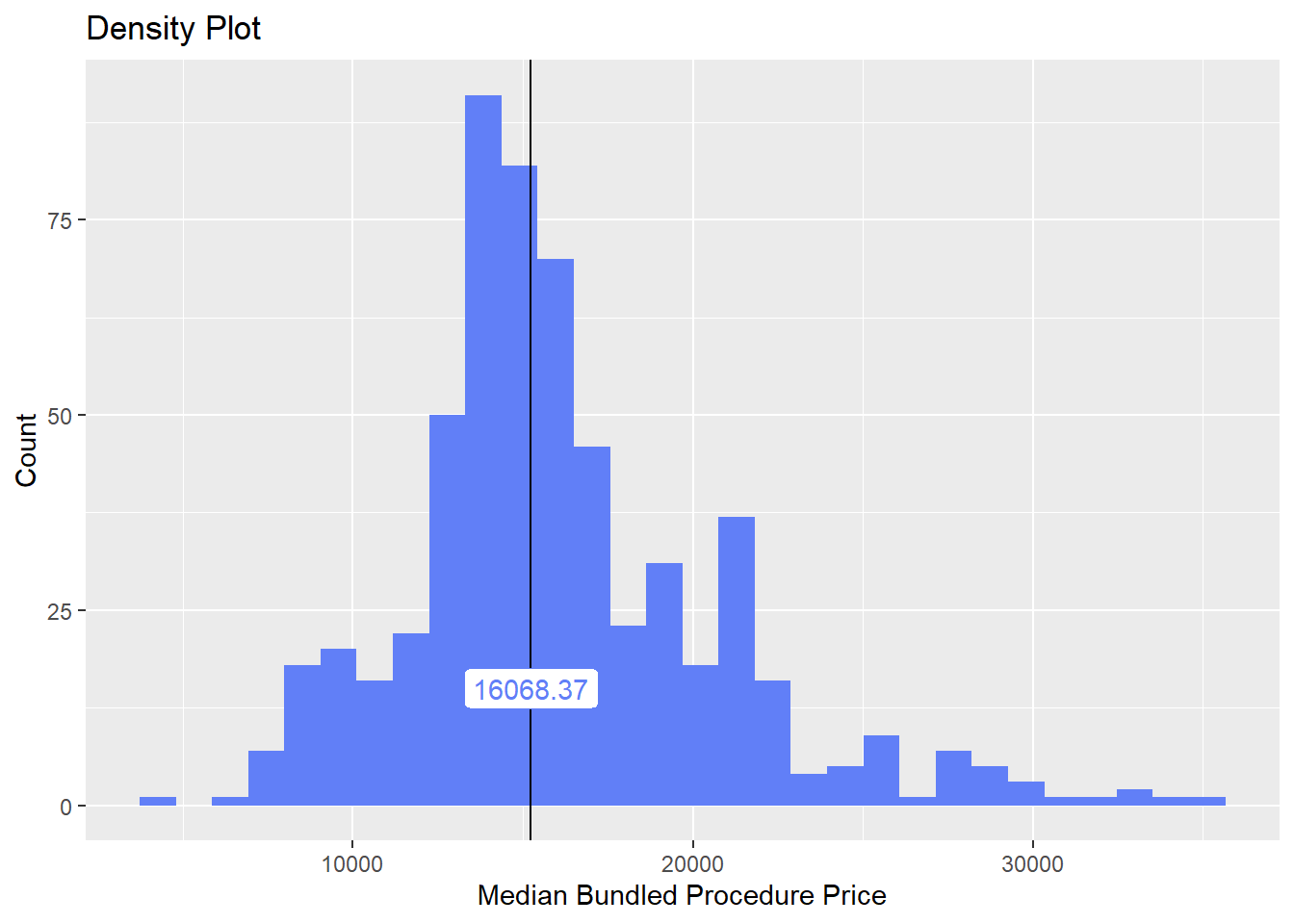
birth_csec_w_anes %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 75 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.110
## 2 medi_bun_t_observ 0.0641
## 3 radi_bun_t_abdom 0.0623
## 4 surg_bun_t_ant 0.0617
## 5 surg_bun_t_antepartum 0.0617
## 6 surg_bun_t_art 0.0617
## 7 surg_bun_t_part 0.0617
## 8 surg_bun_t_tum 0.0617
## 9 path_bun_t_patho 0.061
## 10 path_bun_t_pathologist 0.061
## # ... with 65 more rowsbirth_csec_w_anes <- birth_csec_w_anes[ tp_med > 6000]birth_csec_w_anes_btbv4 <- birth_csec_w_anes %>% btbv4()birth_csec_w_anes_bq <- birth_csec_w_anes_btbv4[,
primary_doctor := pmap(.l=list(doctor_npi1=doctor_npi_str1,
doctor_npi2=doctor_npi_str2,
class_reqs="Obstetrics & Gynecology",
specialization_reqs = "Obstetrics|||Maternal & Fetal Medicine"
),
.f=calculate_primary_doctor)
] %>%
#Filter out any procedures where our doctors fail both criteria.
.[!(primary_doctor %in% c("BOTH_DOC_FAIL_CRIT", "TWO_FIT_ALL_SPECS", "NONE_FIT_SPEC_REQ"))] %>%
.[,primary_doctor_npi := fifelse(primary_doctor==doctor_str1,
doctor_npi_str1,
doctor_npi_str2)] %>%
.[,`:=`(procedure_type=4, procedure_modifier="Caesarean Delivery")]## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "ERIN A.S. CLARK" "ROBERT M. SILVER"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "CHAD C LUNT"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "LAUREN H THEILEN" "MARCELA SMID"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "ROBERT WENDALL LATER"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "BRIAN L WOLSEY"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "LAYNE A SMITH"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "DOUGLAS A ALLEN"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "DOUGLAS A ALLEN"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "CHAD C LUNT"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "MICHAEL L DRAPER"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "ROBERT L ANDRES"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "DOUGLAS A ALLEN"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "ROBERT L ANDRES"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "CARA C HEUSER"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "CHAD C LUNT"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "JOHN DAVID LARAWAY"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)##birth_csec_w_anes_w_sub_hosp_care
we will now look at the density distribution for csection births with anesthesia that also had subsequent hospital care
birth_csec_w_anes_w_sub_hosp_care %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
birth_csec_w_anes_w_sub_hosp_care %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 110 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_cs 0.127
## 2 radi_bun_t_contrast 0.119
## 3 medi_bun_t_immun 0.119
## 4 medi_bun_t_immunization 0.119
## 5 medi_bun_t_vacc 0.119
## 6 medi_bun_t_iiv 0.118
## 7 anes_bun_t_add - on 0.117
## 8 medi_bun_t_clinic 0.112
## 9 medi_bun_t_outpt 0.112
## 10 radi_bun_t_ct 0.111
## # ... with 100 more rowsbirth_csec_w_anes_w_sub_hosp_care <- birth_csec_w_anes_w_sub_hosp_care[ tp_med > 6000]birth_csec_w_anes_w_sub_hosp_care_btbv4 <- birth_csec_w_anes_w_sub_hosp_care %>% btbv4()birth_csec_w_anes_w_sub_hosp_care_bq <- birth_csec_w_anes_w_sub_hosp_care_btbv4[,
primary_doctor := pmap(.l=list(doctor_npi1=doctor_npi_str1,
doctor_npi2=doctor_npi_str2,
class_reqs="Obstetrics & Gynecology",
specialization_reqs = "Obstetrics|||Maternal & Fetal Medicine"
),
.f=calculate_primary_doctor) %>% as.character()
] %>%
#Filter out any procedures where our doctors fail both criteria.
.[!(primary_doctor %in% c("BOTH_DOC_FAIL_CRIT", "TWO_FIT_ALL_SPECS", "NONE_FIT_SPEC_REQ"))] %>%
.[,primary_doctor_npi := fifelse(primary_doctor==doctor_str1,
doctor_npi_str1,
doctor_npi_str2)] %>%
.[,`:=`(procedure_type=5, procedure_modifier="Caesarean Delivery with Subsequent Hospital Care")]## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "ROBERT WENDALL LATER"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "TROY FLINT PORTER"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "CHAD C LUNT"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "ROBERT WENDALL LATER"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "LAYNE A SMITH"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "JHENETTE RENEE LAUDER" "AMY E SULLIVAN"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "TROY FLINT PORTER" "CARA C HEUSER"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "LAYNE A SMITH"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "DONNA DIZON-TOWNSON" "JOHN DAVID LARAWAY"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "TROY FLINT PORTER"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "NATALIE K LOEWEN"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "MICHELLE LYNNE PRECOURT DEBBINK"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "JOHN THOMAS PAAS"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "ROBERT WENDALL LATER"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "GLENN K SCHEMMER"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "JOHN DAVID LARAWAY"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "PAHL BENCH" "BRIAN L WOLSEY"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "JOHN DAVID LARAWAY"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "TROY FLINT PORTER" "DOUGLAS S RICHARDS"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "MICHELLE LYNNE PRECOURT DEBBINK"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "TROY FLINT PORTER"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "GINA B. COX"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## [1] "BRIAN L WOLSEY"
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)
## [1] "multiple meet class req"## Warning in if (!is.na(specialization_reqs)) {: the condition has length > 1 and
## only the first element will be used## character(0)birth_csec_w_anes_w_sub_hosp_care_bq <- birth_csec_w_anes_w_sub_hosp_care_bq[,.(
primary_doctor,
primary_doctor_npi,
most_important_fac ,
most_important_fac_npi,
procedure_type,
procedure_modifier,
tp_med_med,
tp_med_surg,
tp_med_medi,
tp_med_path,
tp_med_radi,
tp_med_anes,
tp_med_faci,
ingest_date = Sys.Date()
)]bq_table_upload(x=procedure_table, values= birth_csec_w_anes_w_sub_hosp_care_bq, create_disposition='CREATE_IF_NEEDED', write_disposition='WRITE_APPEND')