Ingrown Toe Nail
Introduction
In this section we will: * identify which Ingrown Toe Nail procedures were excluded and why * identify which different Ingrown Toe Nail subgroups were created and why
If you have questions or concerns about this data please contact Alexander Nielson (alexnielson@utah.gov)
Load Libraries
Load Libraries
library(data.table)
library(tidyverse)## Warning: replacing previous import 'vctrs::data_frame' by 'tibble::data_frame'
## when loading 'dplyr'library(stringi)
library(ggridges)
library(broom)
library(disk.frame)
library(RecordLinkage)
library(googlesheets4)
library(bigrquery)
library(DBI)
devtools::install_github("utah-osa/hcctools2", upgrade="never" )
library(hcctools2)Establish color palettes
cust_color_pal1 <- c(
"Anesthesia" = "#f3476f",
"Facility" = "#e86a33",
"Medicine Proc" = "#e0a426",
"Pathology" = "#77bf45",
"Radiology" = "#617ff7",
"Surgery" = "#a974ee"
)
cust_color_pal2 <- c(
"TRUE" = "#617ff7",
"FALSE" = "#e0a426"
)
cust_color_pal3 <- c(
"above avg" = "#f3476f",
"avg" = "#77bf45",
"below avg" = "#a974ee"
)
fac_ref_regex <- "(UTAH)|(IHC)|(HOSP)|(HOSPITAL)|(CLINIC)|(ANESTH)|(SCOPY)|(SURG)|(LLC)|(ASSOC)|(MEDIC)|(CENTER)|(ASSOCIATES)|(U OF U)|(HEALTH)|(OLOGY)|(OSCOPY)|(FAMILY)|(VAMC)|(SLC)|(SALT LAKE)|(CITY)|(PROVO)|(OGDEN)|(ENDO )|( VALLEY)|( REGIONAL)|( CTR)|(GRANITE)|( INSTITUTE)|(INSTACARE)|(WASATCH)|(COUNTY)|(PEDIATRIC)|(CORP)|(CENTRAL)|(URGENT)|(CARE)|(UNIV)|(ODYSSEY)|(MOUNTAINSTAR)|( ORTHOPEDIC)|(INSTITUT)|(PARTNERSHIP)|(PHYSICIAN)|(CASTLEVIEW)|(CONSULTING)|(MAGEMENT)|(PRACTICE)|(EMERGENCY)|(SPECIALISTS)|(DIVISION)|(GUT WHISPERER)|(INTERMOUNTAIN)|(OBGYN)"Connect to GCP database
bigrquery::bq_auth(path = 'D:/gcp_keys/healthcare-compare-prod-95b3b7349c32.json')
# set my project ID and dataset name
project_id <- 'healthcare-compare-prod'
dataset_name <- 'healthcare_compare'
con <- dbConnect(
bigrquery::bigquery(),
project = project_id,
dataset = dataset_name,
billing = project_id
)Get NPI table
query <- paste0("SELECT npi, clean_name, osa_group, osa_class, osa_specialization
FROM `healthcare-compare-prod.healthcare_compare.npi_full`")
#bq_project_query(billing, query) # uncomment to determine billing price for above query.
npi_full <- dbGetQuery(con, query) %>%
data.table()get a subset of the NPI providers based upon taxonomy groups
gs4_auth(email="alexnielson@utah.gov")surgery <- read_sheet("https://docs.google.com/spreadsheets/d/1GY8lKwUJuPHtpUl4EOw9eriLUDG9KkNWrMbaSnA5hOU/edit#gid=0",
sheet="major_surgery") %>% as.data.table## Auto-refreshing stale OAuth token.## Reading from "Doctor Types to Keep"## Range "'major_surgery'"surgery <- surgery[is.na(Remove) ] %>% .[["NUCC Classification"]] npi_prov_pair <- npi_full[osa_class %in% surgery] %>%
.[,.(npi=npi,
clean_name = clean_name
)
]Load Data
bun_proc <- disk.frame("full_apcd.df")toe_nail2 <- bun_proc[proc_code_str_sorted %>% stri_detect_regex("11750")]toe_nail2 %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail2 %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 386 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_tum 0.461
## 2 anes_bun_t_anes 0.374
## 3 anes_bun_t_anesth 0.370
## 4 surg_bun_t_blood 0.337
## 5 surg_bun_t_vessel 0.337
## 6 radi_bun_t_elbow 0.337
## 7 faci_bun_t_care 0.337
## 8 faci_bun_t_hospital 0.337
## 9 faci_bun_t_initial 0.337
## 10 surg_bun_t_foot 0.333
## # ... with 376 more rowscols_to_check <- toe_nail2 %>% select_if(is.logical)
tag_frequency <- apply(cols_to_check, 2, function(i) sum(i > 0)) %>% enframe() %>% as.data.table() %>% .[,.(tag=name, freq=value)]
outlier_tag_list <- tag_frequency[freq < 2] %>% .[["tag"]]toe_nail2_clean <- remove_bun_proc_with_rare_tags(outlier_tag_list=outlier_tag_list, df=toe_nail2)## [1] "Start of rare tag removal"
## [1] "nrow:5537 , ncol:1404"
## [1] "nrow:5537"
## [1] "removing: surg_bun_t_a-grft"
## [1] 5536
## [1] "removing: surg_bun_t_abd"
## [1] 5536
## [1] "removing: surg_bun_t_abdom"
## [1] 5536
## [1] "removing: surg_bun_t_abdomen"
## [1] 5536
## [1] "removing: surg_bun_t_ablate"
## [1] 5536
## [1] "removing: surg_bun_t_abortion"
## [1] 5536
## [1] "removing: surg_bun_t_acell"
## [1] 5536
## [1] "removing: surg_bun_t_acellular"
## [1] 5536
## [1] "removing: surg_bun_t_acne"
## [1] 5536
## [1] "removing: surg_bun_t_acute"
## [1] 5536
## [1] "removing: surg_bun_t_adenoid"
## [1] 5536
## [1] "removing: surg_bun_t_adjust"
## [1] 5536
## [1] "removing: surg_bun_t_airway"
## [1] 5536
## [1] "removing: surg_bun_t_anal"
## [1] 5535
## [1] "removing: surg_bun_t_analysis"
## [1] 5535
## [1] "removing: surg_bun_t_aneurysm"
## [1] 5535
## [1] "removing: surg_bun_t_anoscopy"
## [1] 5535
## [1] "removing: surg_bun_t_antepartum"
## [1] 5535
## [1] "removing: surg_bun_t_aorta"
## [1] 5535
## [1] "removing: surg_bun_t_aortic"
## [1] 5535
## [1] "removing: surg_bun_t_aply"
## [1] 5535
## [1] "removing: surg_bun_t_appendectomy"
## [1] 5535
## [1] "removing: surg_bun_t_application"
## [1] 5535
## [1] "removing: surg_bun_t_apply"
## [1] 5535
## [1] "removing: surg_bun_t_approach"
## [1] 5535
## [1] "removing: surg_bun_t_arm"
## [1] 5534
## [1] "removing: surg_bun_t_arterial"
## [1] 5534
## [1] "removing: surg_bun_t_artery"
## [1] 5534
## [1] "removing: surg_bun_t_arthroplasty"
## [1] 5534
## [1] "removing: surg_bun_t_artificial"
## [1] 5534
## [1] "removing: surg_bun_t_augmentation"
## [1] 5534
## [1] "removing: surg_bun_t_autograft"
## [1] 5534
## [1] "removing: surg_bun_t_back"
## [1] 5534
## [1] "removing: surg_bun_t_balloon"
## [1] 5534
## [1] "removing: surg_bun_t_biceps"
## [1] 5534
## [1] "removing: surg_bun_t_bile"
## [1] 5534
## [1] "removing: surg_bun_t_bilobectomy"
## [1] 5534
## [1] "removing: surg_bun_t_bladder"
## [1] 5533
## [1] "removing: surg_bun_t_bleeding"
## [1] 5533
## [1] "removing: surg_bun_t_blockage"
## [1] 5533
## [1] "removing: surg_bun_t_blood"
## [1] 5532
## [1] "removing: surg_bun_t_bowel"
## [1] 5532
## [1] "removing: surg_bun_t_brace"
## [1] 5532
## [1] "removing: surg_bun_t_brain"
## [1] 5532
## [1] "removing: surg_bun_t_breast"
## [1] 5532
## [1] "removing: surg_bun_t_bronchial"
## [1] 5532
## [1] "removing: surg_bun_t_bronchoscopy"
## [1] 5532
## [1] "removing: surg_bun_t_brow"
## [1] 5532
## [1] "removing: surg_bun_t_bulge"
## [1] 5532
## [1] "removing: surg_bun_t_burn"
## [1] 5532
## [1] "removing: surg_bun_t_burr"
## [1] 5532
## [1] "removing: surg_bun_t_buttck"
## [1] 5532
## [1] "removing: surg_bun_t_buttock"
## [1] 5532
## [1] "removing: surg_bun_t_byp"
## [1] 5532
## [1] "removing: surg_bun_t_bypass"
## [1] 5532
## [1] "removing: surg_bun_t_cabg"
## [1] 5532
## [1] "removing: surg_bun_t_canal"
## [1] 5532
## [1] "removing: surg_bun_t_cannula"
## [1] 5532
## [1] "removing: surg_bun_t_capillary"
## [1] 5532
## [1] "removing: surg_bun_t_capsule"
## [1] 5532
## [1] "removing: surg_bun_t_care"
## [1] 5532
## [1] "removing: surg_bun_t_carotid"
## [1] 5532
## [1] "removing: surg_bun_t_cartilage"
## [1] 5532
## [1] "removing: surg_bun_t_cast"
## [1] 5532
## [1] "removing: surg_bun_t_cath"
## [1] 5532
## [1] "removing: surg_bun_t_cavity"
## [1] 5532
## [1] "removing: surg_bun_t_cell"
## [1] 5532
## [1] "removing: surg_bun_t_cervix"
## [1] 5532
## [1] "removing: surg_bun_t_cesarean"
## [1] 5532
## [1] "removing: surg_bun_t_chamber"
## [1] 5532
## [1] "removing: surg_bun_t_change"
## [1] 5532
## [1] "removing: surg_bun_t_chemical"
## [1] 5532
## [1] "removing: surg_bun_t_chemodenerv"
## [1] 5532
## [1] "removing: surg_bun_t_chest"
## [1] 5532
## [1] "removing: surg_bun_t_chin"
## [1] 5532
## [1] "removing: surg_bun_t_cholangiogram"
## [1] 5532
## [1] "removing: surg_bun_t_cholangiopancreatograph"
## [1] 5532
## [1] "removing: surg_bun_t_cholecystoenterostomy"
## [1] 5532
## [1] "removing: surg_bun_t_chronic"
## [1] 5532
## [1] "removing: surg_bun_t_circulation"
## [1] 5532
## [1] "removing: surg_bun_t_circumcision"
## [1] 5532
## [1] "removing: surg_bun_t_clamp"
## [1] 5532
## [1] "removing: surg_bun_t_clavicle"
## [1] 5532
## [1] "removing: surg_bun_t_clearance"
## [1] 5532
## [1] "removing: surg_bun_t_cleft"
## [1] 5532
## [1] "removing: surg_bun_t_clitoris"
## [1] 5532
## [1] "removing: surg_bun_t_cloacal"
## [1] 5532
## [1] "removing: surg_bun_t_closed"
## [1] 5532
## [1] "removing: surg_bun_t_closure"
## [1] 5531
## [1] "removing: surg_bun_t_clot"
## [1] 5531
## [1] "removing: surg_bun_t_collar"
## [1] 5531
## [1] "removing: surg_bun_t_colon"
## [1] 5530
## [1] "removing: surg_bun_t_colonoscopy"
## [1] 5530
## [1] "removing: surg_bun_t_coloproctostomy"
## [1] 5530
## [1] "removing: surg_bun_t_colostomy"
## [1] 5530
## [1] "removing: surg_bun_t_complex"
## [1] 5530
## [1] "removing: surg_bun_t_contriction"
## [1] 5530
## [1] "removing: surg_bun_t_control"
## [1] 5529
## [1] "removing: surg_bun_t_cord"
## [1] 5529
## [1] "removing: surg_bun_t_cornea"
## [1] 5529
## [1] "removing: surg_bun_t_coronary"
## [1] 5529
## [1] "removing: surg_bun_t_coronoid"
## [1] 5529
## [1] "removing: surg_bun_t_cranial"
## [1] 5529
## [1] "removing: surg_bun_t_craniofacial"
## [1] 5529
## [1] "removing: surg_bun_t_cryotherapy"
## [1] 5529
## [1] "removing: surg_bun_t_cuff"
## [1] 5529
## [1] "removing: surg_bun_t_cult"
## [1] 5529
## [1] "removing: surg_bun_t_cysto"
## [1] 5528
## [1] "removing: surg_bun_t_cystometrogram"
## [1] 5528
## [1] "removing: surg_bun_t_cystoscopy"
## [1] 5528
## [1] "removing: surg_bun_t_cystouretero"
## [1] 5528
## [1] "removing: surg_bun_t_deep"
## [1] 5528
## [1] "removing: surg_bun_t_defect"
## [1] 5528
## [1] "removing: surg_bun_t_defib"
## [1] 5528
## [1] "removing: surg_bun_t_degenerated"
## [1] 5528
## [1] "removing: surg_bun_t_delivery"
## [1] 5528
## [1] "removing: surg_bun_t_deplet"
## [1] 5528
## [1] "removing: surg_bun_t_dermabrasion"
## [1] 5528
## [1] "removing: surg_bun_t_design"
## [1] 5528
## [1] "removing: surg_bun_t_destroy"
## [1] 5528
## [1] "removing: surg_bun_t_destruction"
## [1] 5528
## [1] "removing: surg_bun_t_dialysis"
## [1] 5528
## [1] "removing: surg_bun_t_diaphragm"
## [1] 5528
## [1] "removing: surg_bun_t_dilat"
## [1] 5527
## [1] "removing: surg_bun_t_dilation"
## [1] 5527
## [1] "removing: surg_bun_t_disc"
## [1] 5527
## [1] "removing: surg_bun_t_diskectomy"
## [1] 5527
## [1] "removing: surg_bun_t_dissect"
## [1] 5527
## [1] "removing: surg_bun_t_donar"
## [1] 5527
## [1] "removing: surg_bun_t_donor"
## [1] 5527
## [1] "removing: surg_bun_t_draw"
## [1] 5527
## [1] "removing: surg_bun_t_dressing"
## [1] 5527
## [1] "removing: surg_bun_t_drill"
## [1] 5527
## [1] "removing: surg_bun_t_duoden"
## [1] 5527
## [1] "removing: surg_bun_t_ear"
## [1] 5527
## [1] "removing: surg_bun_t_ectopic"
## [1] 5527
## [1] "removing: surg_bun_t_elbow"
## [1] 5526
## [1] "removing: surg_bun_t_electrical"
## [1] 5526
## [1] "removing: surg_bun_t_electrode"
## [1] 5526
## [1] "removing: surg_bun_t_electroejaculation"
## [1] 5526
## [1] "removing: surg_bun_t_electrolysis"
## [1] 5526
## [1] "removing: surg_bun_t_embolize"
## [1] 5526
## [1] "removing: surg_bun_t_embryo"
## [1] 5526
## [1] "removing: surg_bun_t_endovas"
## [1] 5526
## [1] "removing: surg_bun_t_enlarge"
## [1] 5526
## [1] "removing: surg_bun_t_enterectomy"
## [1] 5526
## [1] "removing: surg_bun_t_epiderm"
## [1] 5526
## [1] "removing: surg_bun_t_epididymis"
## [1] 5526
## [1] "removing: surg_bun_t_epiphysis"
## [1] 5526
## [1] "removing: surg_bun_t_episiotomy"
## [1] 5526
## [1] "removing: surg_bun_t_esoph"
## [1] 5526
## [1] "removing: surg_bun_t_esophagoscopy"
## [1] 5526
## [1] "removing: surg_bun_t_esophagus"
## [1] 5526
## [1] "removing: surg_bun_t_exam"
## [1] 5526
## [1] "removing: surg_bun_t_excessive"
## [1] 5526
## [1] "removing: surg_bun_t_excise"
## [1] 5526
## [1] "removing: surg_bun_t_exostosis"
## [1] 5526
## [1] "removing: surg_bun_t_expander"
## [1] 5526
## [1] "removing: surg_bun_t_explora"
## [1] 5526
## [1] "removing: surg_bun_t_explore"
## [1] 5525
## [1] "removing: surg_bun_t_extra"
## [1] 5525
## [1] "removing: surg_bun_t_extremity"
## [1] 5525
## [1] "removing: surg_bun_t_eyelashes"
## [1] 5525
## [1] "removing: surg_bun_t_eyelid"
## [1] 5524
## [1] "removing: surg_bun_t_fallopian"
## [1] 5524
## [1] "removing: surg_bun_t_fat"
## [1] 5524
## [1] "removing: surg_bun_t_femur"
## [1] 5524
## [1] "removing: surg_bun_t_fibula"
## [1] 5524
## [1] "removing: surg_bun_t_fissure"
## [1] 5524
## [1] "removing: surg_bun_t_fistula"
## [1] 5524
## [1] "removing: surg_bun_t_fixation"
## [1] 5524
## [1] "removing: surg_bun_t_flap"
## [1] 5523
## [1] "removing: surg_bun_t_flow"
## [1] 5523
## [1] "removing: surg_bun_t_fluid"
## [1] 5522
## [1] "removing: surg_bun_t_fna"
## [1] 5522
## [1] "removing: surg_bun_t_follow-up"
## [1] 5522
## [1] "removing: surg_bun_t_forearm"
## [1] 5522
## [1] "removing: surg_bun_t_forehead"
## [1] 5522
## [1] "removing: surg_bun_t_free"
## [1] 5522
## [1] "removing: surg_bun_t_frontal"
## [1] 5522
## [1] "removing: surg_bun_t_fundoplasty"
## [1] 5522
## [1] "removing: surg_bun_t_gallbladder"
## [1] 5522
## [1] "removing: surg_bun_t_gastrointestinal"
## [1] 5522
## [1] "removing: surg_bun_t_gastrostomy"
## [1] 5522
## [1] "removing: surg_bun_t_gentialia"
## [1] 5522
## [1] "removing: surg_bun_t_gi"
## [1] 5522
## [1] "removing: surg_bun_t_gland"
## [1] 5522
## [1] "removing: surg_bun_t_glaucoma"
## [1] 5522
## [1] "removing: surg_bun_t_growth"
## [1] 5522
## [1] "removing: surg_bun_t_gum"
## [1] 5522
## [1] "removing: surg_bun_t_hair"
## [1] 5522
## [1] "removing: surg_bun_t_halo"
## [1] 5522
## [1] "removing: surg_bun_t_harvest"
## [1] 5522
## [1] "removing: surg_bun_t_head"
## [1] 5522
## [1] "removing: surg_bun_t_heart"
## [1] 5522
## [1] "removing: surg_bun_t_hematoma"
## [1] 5522
## [1] "removing: surg_bun_t_hemorrhoid"
## [1] 5521
## [1] "removing: surg_bun_t_hemorrhoidopexy"
## [1] 5521
## [1] "removing: surg_bun_t_hernia"
## [1] 5521
## [1] "removing: surg_bun_t_hip"
## [1] 5521
## [1] "removing: surg_bun_t_hole"
## [1] 5521
## [1] "removing: surg_bun_t_hormone"
## [1] 5521
## [1] "removing: surg_bun_t_humer"
## [1] 5521
## [1] "removing: surg_bun_t_hydrocele"
## [1] 5521
## [1] "removing: surg_bun_t_hyst"
## [1] 5521
## [1] "removing: surg_bun_t_hysterectomy"
## [1] 5521
## [1] "removing: surg_bun_t_iliopsoas"
## [1] 5521
## [1] "removing: surg_bun_t_illiac"
## [1] 5521
## [1] "removing: surg_bun_t_imag"
## [1] 5521
## [1] "removing: surg_bun_t_infect"
## [1] 5521
## [1] "removing: surg_bun_t_injection"
## [1] 5520
## [1] "removing: surg_bun_t_inner"
## [1] 5520
## [1] "removing: surg_bun_t_internal"
## [1] 5520
## [1] "removing: surg_bun_t_intestine"
## [1] 5520
## [1] "removing: surg_bun_t_intracorporeal"
## [1] 5520
## [1] "removing: surg_bun_t_intracranial"
## [1] 5520
## [1] "removing: surg_bun_t_intranasal"
## [1] 5520
## [1] "removing: surg_bun_t_inverted"
## [1] 5520
## [1] "removing: surg_bun_t_iris"
## [1] 5520
## [1] "removing: surg_bun_t_jaw"
## [1] 5520
## [1] "removing: surg_bun_t_kidney"
## [1] 5520
## [1] "removing: surg_bun_t_kneecap"
## [1] 5520
## [1] "removing: surg_bun_t_knuckle"
## [1] 5520
## [1] "removing: surg_bun_t_kyphectomy"
## [1] 5520
## [1] "removing: surg_bun_t_laceration"
## [1] 5520
## [1] "removing: surg_bun_t_laminotomy"
## [1] 5520
## [1] "removing: surg_bun_t_lap"
## [1] 5520
## [1] "removing: surg_bun_t_laparoscop"
## [1] 5520
## [1] "removing: surg_bun_t_large"
## [1] 5520
## [1] "removing: surg_bun_t_laryngoscopy"
## [1] 5520
## [1] "removing: surg_bun_t_larynx"
## [1] 5520
## [1] "removing: surg_bun_t_laser"
## [1] 5519
## [1] "removing: surg_bun_t_lavage"
## [1] 5519
## [1] "removing: surg_bun_t_lefort"
## [1] 5519
## [1] "removing: surg_bun_t_lengthen"
## [1] 5519
## [1] "removing: surg_bun_t_ligation"
## [1] 5519
## [1] "removing: surg_bun_t_lining"
## [1] 5519
## [1] "removing: surg_bun_t_lip"
## [1] 5519
## [1] "removing: surg_bun_t_lipectomy"
## [1] 5519
## [1] "removing: surg_bun_t_liver"
## [1] 5519
## [1] "removing: surg_bun_t_lobectomy"
## [1] 5519
## [1] "removing: surg_bun_t_long"
## [1] 5519
## [1] "removing: surg_bun_t_lumbar"
## [1] 5519
## [1] "removing: surg_bun_t_lung"
## [1] 5519
## [1] "removing: surg_bun_t_lymph"
## [1] 5519
## [1] "removing: surg_bun_t_major"
## [1] 5519
## [1] "removing: surg_bun_t_mandible"
## [1] 5519
## [1] "removing: surg_bun_t_marrow"
## [1] 5519
## [1] "removing: surg_bun_t_mast"
## [1] 5519
## [1] "removing: surg_bun_t_mastectomy"
## [1] 5519
## [1] "removing: surg_bun_t_mastoid"
## [1] 5519
## [1] "removing: surg_bun_t_material"
## [1] 5519
## [1] "removing: surg_bun_t_maxilla"
## [1] 5519
## [1] "removing: surg_bun_t_mcp"
## [1] 5519
## [1] "removing: surg_bun_t_mesh"
## [1] 5519
## [1] "removing: surg_bun_t_metacarpal"
## [1] 5519
## [1] "removing: surg_bun_t_microvasc"
## [1] 5519
## [1] "removing: surg_bun_t_middle"
## [1] 5519
## [1] "removing: surg_bun_t_midfoot"
## [1] 5519
## [1] "removing: surg_bun_t_mitral"
## [1] 5519
## [1] "removing: surg_bun_t_mohs"
## [1] 5519
## [1] "removing: surg_bun_t_mouth"
## [1] 5519
## [1] "removing: surg_bun_t_musc"
## [1] 5518
## [1] "removing: surg_bun_t_myelography"
## [1] 5518
## [1] "removing: surg_bun_t_myomectomy"
## [1] 5518
## [1] "removing: surg_bun_t_nasolacrimal"
## [1] 5518
## [1] "removing: surg_bun_t_neck"
## [1] 5518
## [1] "removing: surg_bun_t_needle"
## [1] 5518
## [1] "removing: surg_bun_t_nephrectomy"
## [1] 5518
## [1] "removing: surg_bun_t_neuroelectrode"
## [1] 5518
## [1] "removing: surg_bun_t_neuroendoscopy"
## [1] 5518
## [1] "removing: surg_bun_t_nipple"
## [1] 5518
## [1] "removing: surg_bun_t_node"
## [1] 5518
## [1] "removing: surg_bun_t_nodule"
## [1] 5518
## [1] "removing: surg_bun_t_nonfacial"
## [1] 5518
## [1] "removing: surg_bun_t_nose"
## [1] 5518
## [1] "removing: surg_bun_t_nosebleed"
## [1] 5518
## [1] "removing: surg_bun_t_nuero"
## [1] 5518
## [1] "removing: surg_bun_t_obstetrical"
## [1] 5518
## [1] "removing: surg_bun_t_oral"
## [1] 5518
## [1] "removing: surg_bun_t_orbitocranial"
## [1] 5518
## [1] "removing: surg_bun_t_orchiopexy"
## [1] 5518
## [1] "removing: surg_bun_t_ovarian"
## [1] 5518
## [1] "removing: surg_bun_t_ovary"
## [1] 5518
## [1] "removing: surg_bun_t_pacemaker"
## [1] 5518
## [1] "removing: surg_bun_t_pad"
## [1] 5518
## [1] "removing: surg_bun_t_palate"
## [1] 5518
## [1] "removing: surg_bun_t_palm"
## [1] 5518
## [1] "removing: surg_bun_t_palsy"
## [1] 5518
## [1] "removing: surg_bun_t_pancreas"
## [1] 5518
## [1] "removing: surg_bun_t_pancreatectomy"
## [1] 5518
## [1] "removing: surg_bun_t_pedcle"
## [1] 5518
## [1] "removing: surg_bun_t_pedicle"
## [1] 5518
## [1] "removing: surg_bun_t_peel"
## [1] 5518
## [1] "removing: surg_bun_t_pelvic"
## [1] 5518
## [1] "removing: surg_bun_t_pelvis"
## [1] 5518
## [1] "removing: surg_bun_t_penile"
## [1] 5518
## [1] "removing: surg_bun_t_penis"
## [1] 5517
## [1] "removing: surg_bun_t_percut"
## [1] 5517
## [1] "removing: surg_bun_t_percutaneous"
## [1] 5517
## [1] "removing: surg_bun_t_perineum"
## [1] 5517
## [1] "removing: surg_bun_t_pernineum"
## [1] 5517
## [1] "removing: surg_bun_t_pharynx"
## [1] 5517
## [1] "removing: surg_bun_t_pierce"
## [1] 5517
## [1] "removing: surg_bun_t_pilonidal"
## [1] 5517
## [1] "removing: surg_bun_t_place"
## [1] 5517
## [1] "removing: surg_bun_t_plastic"
## [1] 5517
## [1] "removing: surg_bun_t_pneumonectomy"
## [1] 5517
## [1] "removing: surg_bun_t_polyp"
## [1] 5517
## [1] "removing: surg_bun_t_pouch"
## [1] 5517
## [1] "removing: surg_bun_t_pregnancy"
## [1] 5517
## [1] "removing: surg_bun_t_prep"
## [1] 5517
## [1] "removing: surg_bun_t_prepare"
## [1] 5517
## [1] "removing: surg_bun_t_pressure"
## [1] 5516
## [1] "removing: surg_bun_t_process"
## [1] 5516
## [1] "removing: surg_bun_t_proctosigmoidoscopy"
## [1] 5516
## [1] "removing: surg_bun_t_prolapse"
## [1] 5516
## [1] "removing: surg_bun_t_prostat"
## [1] 5516
## [1] "removing: surg_bun_t_prostate"
## [1] 5516
## [1] "removing: surg_bun_t_prosthesis"
## [1] 5516
## [1] "removing: surg_bun_t_prosthetic"
## [1] 5516
## [1] "removing: surg_bun_t_pulmonary"
## [1] 5516
## [1] "removing: surg_bun_t_punch"
## [1] 5516
## [1] "removing: surg_bun_t_puncture"
## [1] 5516
## [1] "removing: surg_bun_t_pyeloplasty"
## [1] 5516
## [1] "removing: surg_bun_t_radius"
## [1] 5515
## [1] "removing: surg_bun_t_realignment"
## [1] 5515
## [1] "removing: surg_bun_t_rechannel"
## [1] 5515
## [1] "removing: surg_bun_t_reconstruct"
## [1] 5515
## [1] "removing: surg_bun_t_reconstruction"
## [1] 5515
## [1] "removing: surg_bun_t_rectal"
## [1] 5515
## [1] "removing: surg_bun_t_rectum"
## [1] 5515
## [1] "removing: surg_bun_t_reduction"
## [1] 5515
## [1] "removing: surg_bun_t_reinforce"
## [1] 5515
## [1] "removing: surg_bun_t_renal"
## [1] 5515
## [1] "removing: surg_bun_t_replantation"
## [1] 5515
## [1] "removing: surg_bun_t_reset"
## [1] 5515
## [1] "removing: surg_bun_t_retina"
## [1] 5514
## [1] "removing: surg_bun_t_retinopathy"
## [1] 5513
## [1] "removing: surg_bun_t_rib"
## [1] 5513
## [1] "removing: surg_bun_t_ridge"
## [1] 5513
## [1] "removing: surg_bun_t_ring"
## [1] 5513
## [1] "removing: surg_bun_t_roof"
## [1] 5513
## [1] "removing: surg_bun_t_rotator"
## [1] 5513
## [1] "removing: surg_bun_t_routine"
## [1] 5513
## [1] "removing: surg_bun_t_sacrum"
## [1] 5513
## [1] "removing: surg_bun_t_salivary"
## [1] 5513
## [1] "removing: surg_bun_t_scapula"
## [1] 5513
## [1] "removing: surg_bun_t_scar"
## [1] 5513
## [1] "removing: surg_bun_t_scope"
## [1] 5512
## [1] "removing: surg_bun_t_scrotum"
## [1] 5512
## [1] "removing: surg_bun_t_segment"
## [1] 5512
## [1] "removing: surg_bun_t_segmental"
## [1] 5512
## [1] "removing: surg_bun_t_segmentectomy"
## [1] 5512
## [1] "removing: surg_bun_t_septum"
## [1] 5511
## [1] "removing: surg_bun_t_sex"
## [1] 5511
## [1] "removing: surg_bun_t_shorten"
## [1] 5511
## [1] "removing: surg_bun_t_shoulder"
## [1] 5511
## [1] "removing: surg_bun_t_shunt"
## [1] 5511
## [1] "removing: surg_bun_t_sigmoidoscopy"
## [1] 5511
## [1] "removing: surg_bun_t_simple"
## [1] 5511
## [1] "removing: surg_bun_t_sinus"
## [1] 5511
## [1] "removing: surg_bun_t_skeletal"
## [1] 5511
## [1] "removing: surg_bun_t_skull"
## [1] 5511
## [1] "removing: surg_bun_t_sleeve"
## [1] 5511
## [1] "removing: surg_bun_t_slipped"
## [1] 5511
## [1] "removing: surg_bun_t_small"
## [1] 5511
## [1] "removing: surg_bun_t_socket"
## [1] 5511
## [1] "removing: surg_bun_t_soft"
## [1] 5511
## [1] "removing: surg_bun_t_sore"
## [1] 5511
## [1] "removing: surg_bun_t_sphenoid"
## [1] 5511
## [1] "removing: surg_bun_t_sphincter"
## [1] 5511
## [1] "removing: surg_bun_t_spine"
## [1] 5511
## [1] "removing: surg_bun_t_spleen"
## [1] 5511
## [1] "removing: surg_bun_t_splint"
## [1] 5511
## [1] "removing: surg_bun_t_split"
## [1] 5511
## [1] "removing: surg_bun_t_stem"
## [1] 5511
## [1] "removing: surg_bun_t_stenosis"
## [1] 5511
## [1] "removing: surg_bun_t_stent"
## [1] 5511
## [1] "removing: surg_bun_t_sternal"
## [1] 5511
## [1] "removing: surg_bun_t_sternum"
## [1] 5511
## [1] "removing: surg_bun_t_stimulation"
## [1] 5511
## [1] "removing: surg_bun_t_stomach"
## [1] 5511
## [1] "removing: surg_bun_t_stone"
## [1] 5511
## [1] "removing: surg_bun_t_stop"
## [1] 5511
## [1] "removing: surg_bun_t_strapping"
## [1] 5510
## [1] "removing: surg_bun_t_stricture"
## [1] 5510
## [1] "removing: surg_bun_t_styloid"
## [1] 5510
## [1] "removing: surg_bun_t_suction"
## [1] 5510
## [1] "removing: surg_bun_t_superficial"
## [1] 5510
## [1] "removing: surg_bun_t_suspension"
## [1] 5510
## [1] "removing: surg_bun_t_sutures"
## [1] 5510
## [1] "removing: surg_bun_t_sweat"
## [1] 5510
## [1] "removing: surg_bun_t_tags"
## [1] 5510
## [1] "removing: surg_bun_t_tail"
## [1] 5510
## [1] "removing: surg_bun_t_tear"
## [1] 5510
## [1] "removing: surg_bun_t_temporal"
## [1] 5510
## [1] "removing: surg_bun_t_testis"
## [1] 5510
## [1] "removing: surg_bun_t_therapy"
## [1] 5510
## [1] "removing: surg_bun_t_thigh"
## [1] 5510
## [1] "removing: surg_bun_t_thoracoscopy"
## [1] 5510
## [1] "removing: surg_bun_t_thoracotomy"
## [1] 5510
## [1] "removing: surg_bun_t_thorax"
## [1] 5510
## [1] "removing: surg_bun_t_throat"
## [1] 5510
## [1] "removing: surg_bun_t_thrombin"
## [1] 5510
## [1] "removing: surg_bun_t_thrombo"
## [1] 5510
## [1] "removing: surg_bun_t_thymus"
## [1] 5510
## [1] "removing: surg_bun_t_thyroid"
## [1] 5510
## [1] "removing: surg_bun_t_tkr"
## [1] 5510
## [1] "removing: surg_bun_t_tongue"
## [1] 5510
## [1] "removing: surg_bun_t_tonsil"
## [1] 5510
## [1] "removing: surg_bun_t_torsion"
## [1] 5510
## [1] "removing: surg_bun_t_total"
## [1] 5510
## [1] "removing: surg_bun_t_tract"
## [1] 5509
## [1] "removing: surg_bun_t_transcath"
## [1] 5509
## [1] "removing: surg_bun_t_transfer"
## [1] 5509
## [1] "removing: surg_bun_t_transform"
## [1] 5509
## [1] "removing: surg_bun_t_transplant"
## [1] 5508
## [1] "removing: surg_bun_t_triceps"
## [1] 5508
## [1] "removing: surg_bun_t_tricuspid"
## [1] 5508
## [1] "removing: surg_bun_t_trigger"
## [1] 5508
## [1] "removing: surg_bun_t_tube"
## [1] 5508
## [1] "removing: surg_bun_t_tumor"
## [1] 5508
## [1] "removing: surg_bun_t_tunneled"
## [1] 5508
## [1] "removing: surg_bun_t_ulcer"
## [1] 5508
## [1] "removing: surg_bun_t_ulna"
## [1] 5508
## [1] "removing: surg_bun_t_umbil"
## [1] 5508
## [1] "removing: surg_bun_t_unstable"
## [1] 5508
## [1] "removing: surg_bun_t_upper"
## [1] 5508
## [1] "removing: surg_bun_t_ureter"
## [1] 5508
## [1] "removing: surg_bun_t_urethra"
## [1] 5508
## [1] "removing: surg_bun_t_urethrovaginal"
## [1] 5508
## [1] "removing: surg_bun_t_uterus"
## [1] 5508
## [1] "removing: surg_bun_t_uvula"
## [1] 5508
## [1] "removing: surg_bun_t_vag"
## [1] 5507
## [1] "removing: surg_bun_t_vagotomy"
## [1] 5507
## [1] "removing: surg_bun_t_valve"
## [1] 5507
## [1] "removing: surg_bun_t_vascular"
## [1] 5507
## [1] "removing: surg_bun_t_vein"
## [1] 5507
## [1] "removing: surg_bun_t_venipuncture"
## [1] 5507
## [1] "removing: surg_bun_t_ventricular"
## [1] 5507
## [1] "removing: surg_bun_t_vertebra"
## [1] 5507
## [1] "removing: surg_bun_t_vessel"
## [1] 5507
## [1] "removing: surg_bun_t_vulva"
## [1] 5507
## [1] "removing: surg_bun_t_wall"
## [1] 5507
## [1] "removing: surg_bun_t_wax"
## [1] 5507
## [1] "removing: surg_bun_t_web"
## [1] 5507
## [1] "removing: surg_bun_t_wedge"
## [1] 5507
## [1] "removing: surg_bun_t_window"
## [1] 5507
## [1] "removing: surg_bun_t_windpipe"
## [1] 5507
## [1] "removing: surg_bun_t_wound"
## [1] 5507
## [1] "removing: surg_bun_t_wrinkle"
## [1] 5507
## [1] "removing: surg_bun_t_x-ray"
## [1] 5507
## [1] "removing: surg_bun_t_xenograft"
## [1] 5507
## [1] "removing: surg_bun_t_xenogrft"
## [1] 5507
## [1] "removing: surg_bun_t_xgraft"
## [1] 5507
## [1] "removing: medi_bun_t_applicat"
## [1] 5507
## [1] "removing: medi_bun_t_apply"
## [1] 5507
## [1] "removing: medi_bun_t_asses"
## [1] 5507
## [1] "removing: medi_bun_t_benzathine"
## [1] 5507
## [1] "removing: medi_bun_t_candida"
## [1] 5507
## [1] "removing: medi_bun_t_cast"
## [1] 5507
## [1] "removing: medi_bun_t_cathet"
## [1] 5507
## [1] "removing: medi_bun_t_catheterisation"
## [1] 5507
## [1] "removing: medi_bun_t_clostridium"
## [1] 5507
## [1] "removing: medi_bun_t_cochlear"
## [1] 5507
## [1] "removing: medi_bun_t_cold"
## [1] 5506
## [1] "removing: medi_bun_t_consult"
## [1] 5506
## [1] "removing: medi_bun_t_consultation"
## [1] 5506
## [1] "removing: medi_bun_t_crutch"
## [1] 5506
## [1] "removing: medi_bun_t_cyst"
## [1] 5506
## [1] "removing: medi_bun_t_derma"
## [1] 5506
## [1] "removing: medi_bun_t_dermal"
## [1] 5506
## [1] "removing: medi_bun_t_device"
## [1] 5506
## [1] "removing: medi_bun_t_dialys"
## [1] 5506
## [1] "removing: medi_bun_t_discharg"
## [1] 5506
## [1] "removing: medi_bun_t_drain"
## [1] 5506
## [1] "removing: medi_bun_t_draw"
## [1] 5506
## [1] "removing: medi_bun_t_dtap"
## [1] 5506
## [1] "removing: medi_bun_t_dtap-ipv"
## [1] 5506
## [1] "removing: medi_bun_t_endocrine"
## [1] 5506
## [1] "removing: medi_bun_t_epstein-barr"
## [1] 5506
## [1] "removing: medi_bun_t_equipment"
## [1] 5506
## [1] "removing: medi_bun_t_estradiol"
## [1] 5506
## [1] "removing: medi_bun_t_flu"
## [1] 5505
## [1] "removing: medi_bun_t_follow-up"
## [1] 5505
## [1] "removing: medi_bun_t_gastroenterology"
## [1] 5505
## [1] "removing: medi_bun_t_gonorrhoeae"
## [1] 5505
## [1] "removing: medi_bun_t_haloperidol"
## [1] 5505
## [1] "removing: medi_bun_t_hemorrhoid"
## [1] 5505
## [1] "removing: medi_bun_t_hepatitis"
## [1] 5504
## [1] "removing: medi_bun_t_hiv - 2"
## [1] 5504
## [1] "removing: medi_bun_t_hiv - 3"
## [1] 5504
## [1] "removing: medi_bun_t_home"
## [1] 5503
## [1] "removing: medi_bun_t_hormone"
## [1] 5503
## [1] "removing: medi_bun_t_hydration"
## [1] 5503
## [1] "removing: medi_bun_t_immunoassay"
## [1] 5503
## [1] "removing: medi_bun_t_immunohisto"
## [1] 5503
## [1] "removing: medi_bun_t_influenza"
## [1] 5503
## [1] "removing: medi_bun_t_insulin"
## [1] 5503
## [1] "removing: medi_bun_t_intrauterine"
## [1] 5503
## [1] "removing: medi_bun_t_iodine"
## [1] 5503
## [1] "removing: medi_bun_t_mental"
## [1] 5503
## [1] "removing: medi_bun_t_metabolic"
## [1] 5503
## [1] "removing: medi_bun_t_mgmt"
## [1] 5502
## [1] "removing: medi_bun_t_mmr"
## [1] 5501
## [1] "removing: medi_bun_t_monitor"
## [1] 5501
## [1] "removing: medi_bun_t_muscle"
## [1] 5501
## [1] "removing: medi_bun_t_needle"
## [1] 5501
## [1] "removing: medi_bun_t_non-invasive"
## [1] 5501
## [1] "removing: medi_bun_t_non-stress"
## [1] 5501
## [1] "removing: medi_bun_t_normal"
## [1] 5500
## [1] "removing: medi_bun_t_nursing"
## [1] 5500
## [1] "removing: medi_bun_t_nutrition"
## [1] 5499
## [1] "removing: medi_bun_t_observ"
## [1] 5499
## [1] "removing: medi_bun_t_outpatient"
## [1] 5499
## [1] "removing: medi_bun_t_pap"
## [1] 5498
## [1] "removing: medi_bun_t_physical"
## [1] 5498
## [1] "removing: medi_bun_t_portable"
## [1] 5498
## [1] "removing: medi_bun_t_pressure"
## [1] 5498
## [1] "removing: medi_bun_t_psychiat"
## [1] 5498
## [1] "removing: medi_bun_t_psycho"
## [1] 5497
## [1] "removing: medi_bun_t_pulmonary"
## [1] 5497
## [1] "removing: medi_bun_t_rabies"
## [1] 5497
## [1] "removing: medi_bun_t_radiation"
## [1] 5497
## [1] "removing: medi_bun_t_radio"
## [1] 5497
## [1] "removing: medi_bun_t_reeval"
## [1] 5497
## [1] "removing: medi_bun_t_rehab"
## [1] 5497
## [1] "removing: medi_bun_t_remove"
## [1] 5497
## [1] "removing: medi_bun_t_rheumatoid"
## [1] 5497
## [1] "removing: medi_bun_t_rubella"
## [1] 5497
## [1] "removing: medi_bun_t_saline"
## [1] 5497
## [1] "removing: medi_bun_t_sciatic"
## [1] 5497
## [1] "removing: medi_bun_t_serum"
## [1] 5497
## [1] "removing: medi_bun_t_shave"
## [1] 5497
## [1] "removing: medi_bun_t_simple"
## [1] 5497
## [1] "removing: medi_bun_t_sleep"
## [1] 5497
## [1] "removing: medi_bun_t_smear"
## [1] 5497
## [1] "removing: medi_bun_t_soft"
## [1] 5497
## [1] "removing: medi_bun_t_speech"
## [1] 5496
## [1] "removing: medi_bun_t_splint"
## [1] 5495
## [1] "removing: medi_bun_t_stent"
## [1] 5495
## [1] "removing: medi_bun_t_sterile"
## [1] 5494
## [1] "removing: medi_bun_t_stocking"
## [1] 5494
## [1] "removing: medi_bun_t_stone"
## [1] 5494
## [1] "removing: medi_bun_t_stool"
## [1] 5494
## [1] "removing: medi_bun_t_strep"
## [1] 5494
## [1] "removing: medi_bun_t_stress"
## [1] 5493
## [1] "removing: medi_bun_t_subseq"
## [1] 5493
## [1] "removing: medi_bun_t_substitute"
## [1] 5493
## [1] "removing: medi_bun_t_supplies"
## [1] 5493
## [1] "removing: medi_bun_t_suppls"
## [1] 5493
## [1] "removing: medi_bun_t_support"
## [1] 5493
## [1] "removing: medi_bun_t_susceptible"
## [1] 5493
## [1] "removing: medi_bun_t_swallow"
## [1] 5493
## [1] "removing: medi_bun_t_synth"
## [1] 5493
## [1] "removing: medi_bun_t_syphilis"
## [1] 5493
## [1] "removing: medi_bun_t_syringe"
## [1] 5493
## [1] "removing: medi_bun_t_system"
## [1] 5493
## [1] "removing: medi_bun_t_testing"
## [1] 5493
## [1] "removing: medi_bun_t_tests"
## [1] 5492
## [1] "removing: medi_bun_t_tetanus"
## [1] 5492
## [1] "removing: medi_bun_t_thyroxine"
## [1] 5492
## [1] "removing: medi_bun_t_tobacco"
## [1] 5492
## [1] "removing: medi_bun_t_toe"
## [1] 5492
## [1] "removing: medi_bun_t_tolerance"
## [1] 5492
## [1] "removing: medi_bun_t_topical"
## [1] 5492
## [1] "removing: medi_bun_t_toxin"
## [1] 5492
## [1] "removing: medi_bun_t_trach"
## [1] 5492
## [1] "removing: medi_bun_t_training"
## [1] 5491
## [1] "removing: medi_bun_t_transfusion"
## [1] 5491
## [1] "removing: medi_bun_t_treat"
## [1] 5490
## [1] "removing: medi_bun_t_typhoid"
## [1] 5490
## [1] "removing: medi_bun_t_urinalysis"
## [1] 5490
## [1] "removing: medi_bun_t_urinary"
## [1] 5490
## [1] "removing: medi_bun_t_urine"
## [1] 5490
## [1] "removing: medi_bun_t_uterine"
## [1] 5490
## [1] "removing: medi_bun_t_valve"
## [1] 5490
## [1] "removing: medi_bun_t_vein"
## [1] 5490
## [1] "removing: medi_bun_t_virus"
## [1] 5490
## [1] "removing: medi_bun_t_visual"
## [1] 5489
## [1] "removing: medi_bun_t_wbc"
## [1] 5489
## [1] "removing: medi_bun_t_wheelchair"
## [1] 5489
## [1] "removing: medi_bun_t_wheezing"
## [1] 5488
## [1] "removing: medi_bun_t_wire"
## [1] 5488
## [1] "removing: medi_bun_t_with"
## [1] 5487
## [1] "removing: medi_bun_t_without"
## [1] 5487
## [1] "removing: medi_bun_t_wkend"
## [1] 5487
## [1] "removing: medi_bun_t_hzv"
## [1] 5486
## [1] "removing: medi_bun_t_xgraft"
## [1] 5486
## [1] "removing: medi_bun_t_hepb"
## [1] 5486
## [1] "removing: medi_bun_t_ppsv23"
## [1] 5486
## [1] "removing: medi_bun_t_var_vaccine_live_subq"
## [1] 5485
## [1] "removing: radi_bun_t_abdom"
## [1] 5485
## [1] "removing: radi_bun_t_adrenal"
## [1] 5485
## [1] "removing: radi_bun_t_angiography"
## [1] 5485
## [1] "removing: radi_bun_t_aorta"
## [1] 5485
## [1] "removing: radi_bun_t_area"
## [1] 5485
## [1] "removing: radi_bun_t_arm"
## [1] 5485
## [1] "removing: radi_bun_t_artery"
## [1] 5485
## [1] "removing: radi_bun_t_bile"
## [1] 5485
## [1] "removing: radi_bun_t_bladder"
## [1] 5485
## [1] "removing: radi_bun_t_blade"
## [1] 5485
## [1] "removing: radi_bun_t_blockage"
## [1] 5485
## [1] "removing: radi_bun_t_body"
## [1] 5485
## [1] "removing: radi_bun_t_brain"
## [1] 5484
## [1] "removing: radi_bun_t_cardiac"
## [1] 5484
## [1] "removing: radi_bun_t_cast"
## [1] 5484
## [1] "removing: radi_bun_t_complex"
## [1] 5484
## [1] "removing: radi_bun_t_contrast"
## [1] 5484
## [1] "removing: radi_bun_t_cranial"
## [1] 5484
## [1] "removing: radi_bun_t_ct_scan"
## [1] 5484
## [1] "removing: radi_bun_t_delivery"
## [1] 5483
## [1] "removing: radi_bun_t_diagnostic"
## [1] 5483
## [1] "removing: radi_bun_t_dislocation"
## [1] 5483
## [1] "removing: radi_bun_t_duct"
## [1] 5483
## [1] "removing: radi_bun_t_elbow"
## [1] 5483
## [1] "removing: radi_bun_t_epidurograpy"
## [1] 5483
## [1] "removing: radi_bun_t_evaluation"
## [1] 5483
## [1] "removing: radi_bun_t_eye"
## [1] 5483
## [1] "removing: radi_bun_t_femur"
## [1] 5483
## [1] "removing: radi_bun_t_fetal"
## [1] 5483
## [1] "removing: radi_bun_t_fistula"
## [1] 5483
## [1] "removing: radi_bun_t_fmri"
## [1] 5483
## [1] "removing: radi_bun_t_forearm"
## [1] 5483
## [1] "removing: radi_bun_t_fracture"
## [1] 5483
## [1] "removing: radi_bun_t_gadobutrol"
## [1] 5483
## [1] "removing: radi_bun_t_gastric"
## [1] 5483
## [1] "removing: radi_bun_t_general"
## [1] 5483
## [1] "removing: radi_bun_t_gential"
## [1] 5483
## [1] "removing: radi_bun_t_gestation"
## [1] 5483
## [1] "removing: radi_bun_t_gi_tract"
## [1] 5483
## [1] "removing: radi_bun_t_glands"
## [1] 5483
## [1] "removing: radi_bun_t_heart"
## [1] 5483
## [1] "removing: radi_bun_t_heel"
## [1] 5482
## [1] "removing: radi_bun_t_humerus"
## [1] 5482
## [1] "removing: radi_bun_t_hyperthermia"
## [1] 5482
## [1] "removing: radi_bun_t_imag"
## [1] 5482
## [1] "removing: radi_bun_t_image"
## [1] 5482
## [1] "removing: radi_bun_t_imaging"
## [1] 5482
## [1] "removing: radi_bun_t_infant"
## [1] 5482
## [1] "removing: radi_bun_t_interpret"
## [1] 5482
## [1] "removing: radi_bun_t_jaw"
## [1] 5482
## [1] "removing: radi_bun_t_kidney"
## [1] 5482
## [1] "removing: radi_bun_t_leg"
## [1] 5482
## [1] "removing: radi_bun_t_lesion"
## [1] 5482
## [1] "removing: radi_bun_t_liver"
## [1] 5482
## [1] "removing: radi_bun_t_lower"
## [1] 5481
## [1] "removing: radi_bun_t_lung"
## [1] 5481
## [1] "removing: radi_bun_t_mammogra"
## [1] 5481
## [1] "removing: radi_bun_t_management"
## [1] 5481
## [1] "removing: radi_bun_t_marrow"
## [1] 5481
## [1] "removing: radi_bun_t_mastoids"
## [1] 5481
## [1] "removing: radi_bun_t_mra"
## [1] 5481
## [1] "removing: radi_bun_t_myelography"
## [1] 5481
## [1] "removing: radi_bun_t_nuclear"
## [1] 5481
## [1] "removing: radi_bun_t_oncology"
## [1] 5481
## [1] "removing: radi_bun_t_pancreas"
## [1] 5481
## [1] "removing: radi_bun_t_penis"
## [1] 5481
## [1] "removing: radi_bun_t_pet"
## [1] 5481
## [1] "removing: radi_bun_t_pituitary"
## [1] 5481
## [1] "removing: radi_bun_t_plan"
## [1] 5481
## [1] "removing: radi_bun_t_plasma"
## [1] 5481
## [1] "removing: radi_bun_t_radiation"
## [1] 5481
## [1] "removing: radi_bun_t_radio"
## [1] 5481
## [1] "removing: radi_bun_t_repair"
## [1] 5481
## [1] "removing: radi_bun_t_rib"
## [1] 5481
## [1] "removing: radi_bun_t_scan"
## [1] 5481
## [1] "removing: radi_bun_t_sinuses"
## [1] 5481
## [1] "removing: radi_bun_t_skull"
## [1] 5481
## [1] "removing: radi_bun_t_socket"
## [1] 5481
## [1] "removing: radi_bun_t_speech"
## [1] 5481
## [1] "removing: radi_bun_t_spleen"
## [1] 5481
## [1] "removing: radi_bun_t_tear"
## [1] 5481
## [1] "removing: radi_bun_t_teeth"
## [1] 5481
## [1] "removing: radi_bun_t_therapy"
## [1] 5481
## [1] "removing: radi_bun_t_thigh"
## [1] 5481
## [1] "removing: radi_bun_t_thorax"
## [1] 5481
## [1] "removing: radi_bun_t_throat"
## [1] 5481
## [1] "removing: radi_bun_t_thrombosis"
## [1] 5481
## [1] "removing: radi_bun_t_thyroid"
## [1] 5481
## [1] "removing: radi_bun_t_tract"
## [1] 5481
## [1] "removing: radi_bun_t_treatment"
## [1] 5481
## [1] "removing: radi_bun_t_trunk"
## [1] 5481
## [1] "removing: radi_bun_t_tumor"
## [1] 5481
## [1] "removing: radi_bun_t_upper"
## [1] 5481
## [1] "removing: radi_bun_t_urethra"
## [1] 5481
## [1] "removing: radi_bun_t_urinary"
## [1] 5481
## [1] "removing: radi_bun_t_vein"
## [1] 5481
## [1] "removing: radi_bun_t_ventilation"
## [1] 5481
## [1] "removing: radi_bun_t_veous"
## [1] 5481
## [1] "removing: radi_bun_t_vessel"
## [1] 5481
## [1] "removing: radi_bun_t_with_dye"
## [1] 5481
## [1] "removing: radi_bun_t_xray"
## [1] 5480
## [1] "removing: radi_bun_t_xtremity"
## [1] 5480
## [1] "removing: path_bun_t_acet"
## [1] 5479
## [1] "removing: path_bun_t_acute"
## [1] 5479
## [1] "removing: path_bun_t_alkaline"
## [1] 5479
## [1] "removing: path_bun_t_allerg"
## [1] 5479
## [1] "removing: path_bun_t_amikacin"
## [1] 5479
## [1] "removing: path_bun_t_amitripytline"
## [1] 5479
## [1] "removing: path_bun_t_ammonia"
## [1] 5479
## [1] "removing: path_bun_t_amniotic"
## [1] 5479
## [1] "removing: path_bun_t_anabolic"
## [1] 5479
## [1] "removing: path_bun_t_analgesics"
## [1] 5479
## [1] "removing: path_bun_t_analysis"
## [1] 5478
## [1] "removing: path_bun_t_antidepressant"
## [1] 5478
## [1] "removing: path_bun_t_antiepileptics"
## [1] 5478
## [1] "removing: path_bun_t_antipsychotic"
## [1] 5478
## [1] "removing: path_bun_t_aortic"
## [1] 5478
## [1] "removing: path_bun_t_autopsy"
## [1] 5478
## [1] "removing: path_bun_t_barbiturate"
## [1] 5478
## [1] "removing: path_bun_t_benzo"
## [1] 5478
## [1] "removing: path_bun_t_bile"
## [1] 5478
## [1] "removing: path_bun_t_bilirubin"
## [1] 5477
## [1] "removing: path_bun_t_biopsy"
## [1] 5477
## [1] "removing: path_bun_t_bleeding"
## [1] 5477
## [1] "removing: path_bun_t_cadmium"
## [1] 5477
## [1] "removing: path_bun_t_calcium"
## [1] 5477
## [1] "removing: path_bun_t_cannabinoids"
## [1] 5477
## [1] "removing: path_bun_t_carbamazepine"
## [1] 5477
## [1] "removing: path_bun_t_carbon"
## [1] 5477
## [1] "removing: path_bun_t_catecholamine"
## [1] 5477
## [1] "removing: path_bun_t_chlamydia"
## [1] 5477
## [1] "removing: path_bun_t_chloride"
## [1] 5477
## [1] "removing: path_bun_t_chromosome"
## [1] 5477
## [1] "removing: path_bun_t_chromotograph"
## [1] 5477
## [1] "removing: path_bun_t_class"
## [1] 5477
## [1] "removing: path_bun_t_clot"
## [1] 5477
## [1] "removing: path_bun_t_clozapine"
## [1] 5477
## [1] "removing: path_bun_t_coagulation"
## [1] 5477
## [1] "removing: path_bun_t_cocaine"
## [1] 5477
## [1] "removing: path_bun_t_cryopreservd"
## [1] 5477
## [1] "removing: path_bun_t_cyanide"
## [1] 5477
## [1] "removing: path_bun_t_cyclosporine"
## [1] 5477
## [1] "removing: path_bun_t_cystines"
## [1] 5477
## [1] "removing: path_bun_t_cytopathol"
## [1] 5477
## [1] "removing: path_bun_t_dehydroepiandrosterone"
## [1] 5477
## [1] "removing: path_bun_t_desipramine"
## [1] 5477
## [1] "removing: path_bun_t_digoxin"
## [1] 5477
## [1] "removing: path_bun_t_dihydro"
## [1] 5477
## [1] "removing: path_bun_t_dilation"
## [1] 5477
## [1] "removing: path_bun_t_dioxide"
## [1] 5477
## [1] "removing: path_bun_t_dipropylacetic"
## [1] 5477
## [1] "removing: path_bun_t_disease"
## [1] 5477
## [1] "removing: path_bun_t_draw"
## [1] 5477
## [1] "removing: path_bun_t_dysfunction"
## [1] 5477
## [1] "removing: path_bun_t_embryo"
## [1] 5477
## [1] "removing: path_bun_t_encephalitis"
## [1] 5477
## [1] "removing: path_bun_t_endocrine"
## [1] 5477
## [1] "removing: path_bun_t_epstein-barr"
## [1] 5477
## [1] "removing: path_bun_t_estrogen"
## [1] 5477
## [1] "removing: path_bun_t_ethanol"
## [1] 5477
## [1] "removing: path_bun_t_evaluation"
## [1] 5477
## [1] "removing: path_bun_t_fentanyl"
## [1] 5477
## [1] "removing: path_bun_t_fetal"
## [1] 5477
## [1] "removing: path_bun_t_fibrinogen"
## [1] 5477
## [1] "removing: path_bun_t_fibronlytic"
## [1] 5477
## [1] "removing: path_bun_t_flouride"
## [1] 5477
## [1] "removing: path_bun_t_galacto"
## [1] 5477
## [1] "removing: path_bun_t_gases"
## [1] 5477
## [1] "removing: path_bun_t_gastrin"
## [1] 5477
## [1] "removing: path_bun_t_genome"
## [1] 5477
## [1] "removing: path_bun_t_genotpying"
## [1] 5477
## [1] "removing: path_bun_t_glucagon"
## [1] 5477
## [1] "removing: path_bun_t_growth"
## [1] 5477
## [1] "removing: path_bun_t_gynecologic"
## [1] 5477
## [1] "removing: path_bun_t_haloperidol"
## [1] 5477
## [1] "removing: path_bun_t_hearing"
## [1] 5477
## [1] "removing: path_bun_t_heavy"
## [1] 5477
## [1] "removing: path_bun_t_heinz"
## [1] 5477
## [1] "removing: path_bun_t_hematocrit"
## [1] 5477
## [1] "removing: path_bun_t_heridatary"
## [1] 5477
## [1] "removing: path_bun_t_heroin"
## [1] 5477
## [1] "removing: path_bun_t_herpes"
## [1] 5477
## [1] "removing: path_bun_t_hold"
## [1] 5477
## [1] "removing: path_bun_t_hpa"
## [1] 5477
## [1] "removing: path_bun_t_hsv"
## [1] 5477
## [1] "removing: path_bun_t_imipramine"
## [1] 5477
## [1] "removing: path_bun_t_immunoassay"
## [1] 5477
## [1] "removing: path_bun_t_influenza"
## [1] 5476
## [1] "removing: path_bun_t_insulin"
## [1] 5475
## [1] "removing: path_bun_t_iodine"
## [1] 5475
## [1] "removing: path_bun_t_lab"
## [1] 5475
## [1] "removing: path_bun_t_lamotrigine"
## [1] 5475
## [1] "removing: path_bun_t_levetiracetam"
## [1] 5475
## [1] "removing: path_bun_t_lidocaine"
## [1] 5475
## [1] "removing: path_bun_t_lithium"
## [1] 5475
## [1] "removing: path_bun_t_loss"
## [1] 5475
## [1] "removing: path_bun_t_low"
## [1] 5475
## [1] "removing: path_bun_t_lung"
## [1] 5475
## [1] "removing: path_bun_t_lyme"
## [1] 5475
## [1] "removing: path_bun_t_lympg"
## [1] 5475
## [1] "removing: path_bun_t_malaria"
## [1] 5475
## [1] "removing: path_bun_t_mercury"
## [1] 5475
## [1] "removing: path_bun_t_metal"
## [1] 5475
## [1] "removing: path_bun_t_microscop"
## [1] 5474
## [1] "removing: path_bun_t_microscopic"
## [1] 5474
## [1] "removing: path_bun_t_microscopy"
## [1] 5474
## [1] "removing: path_bun_t_mitocondrial"
## [1] 5474
## [1] "removing: path_bun_t_mopath"
## [1] 5474
## [1] "removing: path_bun_t_mumps"
## [1] 5474
## [1] "removing: path_bun_t_mycophenolate"
## [1] 5474
## [1] "removing: path_bun_t_nicotine"
## [1] 5474
## [1] "removing: path_bun_t_nile"
## [1] 5474
## [1] "removing: path_bun_t_non-opioid"
## [1] 5474
## [1] "removing: path_bun_t_nonantibody"
## [1] 5474
## [1] "removing: path_bun_t_nucleic"
## [1] 5474
## [1] "removing: path_bun_t_obstretric"
## [1] 5474
## [1] "removing: path_bun_t_occult"
## [1] 5474
## [1] "removing: path_bun_t_onco"
## [1] 5474
## [1] "removing: path_bun_t_oocyte"
## [1] 5474
## [1] "removing: path_bun_t_opioid"
## [1] 5474
## [1] "removing: path_bun_t_ovulation"
## [1] 5474
## [1] "removing: path_bun_t_oxycodone"
## [1] 5474
## [1] "removing: path_bun_t_paper"
## [1] 5474
## [1] "removing: path_bun_t_period"
## [1] 5474
## [1] "removing: path_bun_t_phenobarbital"
## [1] 5474
## [1] "removing: path_bun_t_phenytoin"
## [1] 5474
## [1] "removing: path_bun_t_phosphatase"
## [1] 5474
## [1] "removing: path_bun_t_pituitary"
## [1] 5474
## [1] "removing: path_bun_t_plasminogen"
## [1] 5474
## [1] "removing: path_bun_t_poliovirus"
## [1] 5474
## [1] "removing: path_bun_t_pregabalin"
## [1] 5474
## [1] "removing: path_bun_t_primidone"
## [1] 5474
## [1] "removing: path_bun_t_procainamide"
## [1] 5474
## [1] "removing: path_bun_t_procedure"
## [1] 5474
## [1] "removing: path_bun_t_prophobulinogen"
## [1] 5474
## [1] "removing: path_bun_t_prophyrins"
## [1] 5474
## [1] "removing: path_bun_t_propoxyphene"
## [1] 5474
## [1] "removing: path_bun_t_prostate"
## [1] 5474
## [1] "removing: path_bun_t_quinide"
## [1] 5474
## [1] "removing: path_bun_t_quinine"
## [1] 5474
## [1] "removing: path_bun_t_reagent"
## [1] 5474
## [1] "removing: path_bun_t_renin"
## [1] 5474
## [1] "removing: path_bun_t_retinal"
## [1] 5474
## [1] "removing: path_bun_t_rickettsia"
## [1] 5474
## [1] "removing: path_bun_t_rotovirus"
## [1] 5474
## [1] "removing: path_bun_t_rubella"
## [1] 5474
## [1] "removing: path_bun_t_salicylate"
## [1] 5474
## [1] "removing: path_bun_t_salmonella"
## [1] 5474
## [1] "removing: path_bun_t_scan"
## [1] 5474
## [1] "removing: path_bun_t_screen"
## [1] 5473
## [1] "removing: path_bun_t_sedative"
## [1] 5473
## [1] "removing: path_bun_t_semen"
## [1] 5472
## [1] "removing: path_bun_t_sequence"
## [1] 5472
## [1] "removing: path_bun_t_serotonin"
## [1] 5472
## [1] "removing: path_bun_t_silica"
## [1] 5472
## [1] "removing: path_bun_t_sirolimus"
## [1] 5471
## [1] "removing: path_bun_t_skin"
## [1] 5471
## [1] "removing: path_bun_t_slenium"
## [1] 5471
## [1] "removing: path_bun_t_specimen"
## [1] 5470
## [1] "removing: path_bun_t_sperm"
## [1] 5470
## [1] "removing: path_bun_t_steroid"
## [1] 5470
## [1] "removing: path_bun_t_stimulation"
## [1] 5470
## [1] "removing: path_bun_t_syphilis"
## [1] 5469
## [1] "removing: path_bun_t_syringe"
## [1] 5469
## [1] "removing: path_bun_t_tacrolims"
## [1] 5469
## [1] "removing: path_bun_t_thawing"
## [1] 5469
## [1] "removing: path_bun_t_theophylline"
## [1] 5469
## [1] "removing: path_bun_t_tobramycin"
## [1] 5469
## [1] "removing: path_bun_t_tolerance"
## [1] 5469
## [1] "removing: path_bun_t_toxoplasma"
## [1] 5469
## [1] "removing: path_bun_t_tpiramate"
## [1] 5469
## [1] "removing: path_bun_t_transfusion"
## [1] 5469
## [1] "removing: path_bun_t_tumor"
## [1] 5469
## [1] "removing: path_bun_t_typing"
## [1] 5469
## [1] "removing: path_bun_t_vancomycin"
## [1] 5469
## [1] "removing: path_bun_t_vein"
## [1] 5469
## [1] "removing: path_bun_t_venipuncture"
## [1] 5469
## [1] "removing: path_bun_t_west"
## [1] 5469
## [1] "removing: path_bun_t_with_scope"
## [1] 5469
## [1] "removing: path_bun_t_zonisamide"
## [1] 5469
## [1] "removing: anes_bun_t_abdom"
## [1] 5469
## [1] "removing: anes_bun_t_add - on"
## [1] 5469
## [1] "removing: anes_bun_t_adrenal"
## [1] 5469
## [1] "removing: anes_bun_t_amputation"
## [1] 5469
## [1] "removing: anes_bun_t_analg"
## [1] 5469
## [1] "removing: anes_bun_t_area"
## [1] 5468
## [1] "removing: anes_bun_t_arest"
## [1] 5468
## [1] "removing: anes_bun_t_arteriography"
## [1] 5468
## [1] "removing: anes_bun_t_artery"
## [1] 5468
## [1] "removing: anes_bun_t_biopsy"
## [1] 5468
## [1] "removing: anes_bun_t_bladder"
## [1] 5468
## [1] "removing: anes_bun_t_blood"
## [1] 5468
## [1] "removing: anes_bun_t_breast"
## [1] 5468
## [1] "removing: anes_bun_t_bronchi"
## [1] 5468
## [1] "removing: anes_bun_t_burn"
## [1] 5468
## [1] "removing: anes_bun_t_casting"
## [1] 5468
## [1] "removing: anes_bun_t_cervix"
## [1] 5468
## [1] "removing: anes_bun_t_chemonucleolysis"
## [1] 5468
## [1] "removing: anes_bun_t_chest"
## [1] 5468
## [1] "removing: anes_bun_t_cleft"
## [1] 5468
## [1] "removing: anes_bun_t_cord"
## [1] 5468
## [1] "removing: anes_bun_t_corneal"
## [1] 5468
## [1] "removing: anes_bun_t_cs"
## [1] 5468
## [1] "removing: anes_bun_t_debride"
## [1] 5468
## [1] "removing: anes_bun_t_deliver"
## [1] 5468
## [1] "removing: anes_bun_t_drainage"
## [1] 5468
## [1] "removing: anes_bun_t_duct"
## [1] 5468
## [1] "removing: anes_bun_t_ear"
## [1] 5468
## [1] "removing: anes_bun_t_elbow"
## [1] 5468
## [1] "removing: anes_bun_t_electroshock"
## [1] 5468
## [1] "removing: anes_bun_t_embolectomy"
## [1] 5468
## [1] "removing: anes_bun_t_endoscopy"
## [1] 5468
## [1] "removing: anes_bun_t_epidural"
## [1] 5468
## [1] "removing: anes_bun_t_esophag"
## [1] 5468
## [1] "removing: anes_bun_t_eyelid"
## [1] 5468
## [1] "removing: anes_bun_t_facial"
## [1] 5468
## [1] "removing: anes_bun_t_fat"
## [1] 5468
## [1] "removing: anes_bun_t_femoral"
## [1] 5468
## [1] "removing: anes_bun_t_fentanyl"
## [1] 5468
## [1] "removing: anes_bun_t_genitalia"
## [1] 5468
## [1] "removing: anes_bun_t_glands"
## [1] 5468
## [1] "removing: anes_bun_t_head"
## [1] 5467
## [1] "removing: anes_bun_t_hernia"
## [1] 5467
## [1] "removing: anes_bun_t_humer"
## [1] 5467
## [1] "removing: anes_bun_t_hydromorphone"
## [1] 5467
## [1] "removing: anes_bun_t_hysterectomy"
## [1] 5467
## [1] "removing: anes_bun_t_image"
## [1] 5467
## [1] "removing: anes_bun_t_insertion"
## [1] 5467
## [1] "removing: anes_bun_t_intracranial"
## [1] 5467
## [1] "removing: anes_bun_t_iredectomy"
## [1] 5467
## [1] "removing: anes_bun_t_joint"
## [1] 5466
## [1] "removing: anes_bun_t_ketorolac"
## [1] 5466
## [1] "removing: anes_bun_t_kidney"
## [1] 5466
## [1] "removing: anes_bun_t_larynx"
## [1] 5466
## [1] "removing: anes_bun_t_layer"
## [1] 5466
## [1] "removing: anes_bun_t_lip"
## [1] 5466
## [1] "removing: anes_bun_t_liver"
## [1] 5466
## [1] "removing: anes_bun_t_local"
## [1] 5466
## [1] "removing: anes_bun_t_lumbar"
## [1] 5466
## [1] "removing: anes_bun_t_lung"
## [1] 5466
## [1] "removing: anes_bun_t_major"
## [1] 5466
## [1] "removing: anes_bun_t_manipulation"
## [1] 5466
## [1] "removing: anes_bun_t_midazolam"
## [1] 5466
## [1] "removing: anes_bun_t_mod_sed"
## [1] 5465
## [1] "removing: anes_bun_t_neck"
## [1] 5465
## [1] "removing: anes_bun_t_nerve"
## [1] 5465
## [1] "removing: anes_bun_t_open"
## [1] 5465
## [1] "removing: anes_bun_t_organ"
## [1] 5465
## [1] "removing: anes_bun_t_pacemaker"
## [1] 5465
## [1] "removing: anes_bun_t_palate"
## [1] 5465
## [1] "removing: anes_bun_t_pelvic"
## [1] 5465
## [1] "removing: anes_bun_t_pelvis"
## [1] 5465
## [1] "removing: anes_bun_t_penis"
## [1] 5465
## [1] "removing: anes_bun_t_percutaneous"
## [1] 5465
## [1] "removing: anes_bun_t_plastic"
## [1] 5465
## [1] "removing: anes_bun_t_propofol"
## [1] 5465
## [1] "removing: anes_bun_t_prostate"
## [1] 5465
## [1] "removing: anes_bun_t_pump"
## [1] 5465
## [1] "removing: anes_bun_t_puncture"
## [1] 5465
## [1] "removing: anes_bun_t_radical"
## [1] 5465
## [1] "removing: anes_bun_t_release"
## [1] 5465
## [1] "removing: anes_bun_t_removal"
## [1] 5465
## [1] "removing: anes_bun_t_repair"
## [1] 5465
## [1] "removing: anes_bun_t_replace"
## [1] 5465
## [1] "removing: anes_bun_t_rib"
## [1] 5465
## [1] "removing: anes_bun_t_sedation"
## [1] 5465
## [1] "removing: anes_bun_t_shoulder"
## [1] 5465
## [1] "removing: anes_bun_t_skull"
## [1] 5465
## [1] "removing: anes_bun_t_sperm"
## [1] 5465
## [1] "removing: anes_bun_t_spine"
## [1] 5465
## [1] "removing: anes_bun_t_spit"
## [1] 5465
## [1] "removing: anes_bun_t_sternal"
## [1] 5465
## [1] "removing: anes_bun_t_stone"
## [1] 5465
## [1] "removing: anes_bun_t_tendon"
## [1] 5465
## [1] "removing: anes_bun_t_testis"
## [1] 5465
## [1] "removing: anes_bun_t_thigh"
## [1] 5465
## [1] "removing: anes_bun_t_thyriod"
## [1] 5465
## [1] "removing: anes_bun_t_trachea"
## [1] 5465
## [1] "removing: anes_bun_t_transplant"
## [1] 5465
## [1] "removing: anes_bun_t_tromethamine"
## [1] 5465
## [1] "removing: anes_bun_t_tubal"
## [1] 5465
## [1] "removing: anes_bun_t_tympanotomy"
## [1] 5465
## [1] "removing: anes_bun_t_upper"
## [1] 5465
## [1] "removing: anes_bun_t_ureter"
## [1] 5465
## [1] "removing: anes_bun_t_urethra"
## [1] 5465
## [1] "removing: anes_bun_t_vag"
## [1] 5465
## [1] "removing: anes_bun_t_vein"
## [1] 5465
## [1] "removing: anes_bun_t_ventilation"
## [1] 5465
## [1] "removing: anes_bun_t_vesectomy"
## [1] 5465
## [1] "removing: anes_bun_t_vessel"
## [1] 5465
## [1] "removing: anes_bun_t_viteroretinal"
## [1] 5465
## [1] "removing: anes_bun_t_wall"
## [1] 5465
## [1] "removing: anes_bun_t_wrist"
## [1] 5465
## [1] "removing: faci_bun_t_advanced"
## [1] 5465
## [1] "removing: faci_bun_t_analysis"
## [1] 5465
## [1] "removing: faci_bun_t_annual"
## [1] 5465
## [1] "removing: faci_bun_t_assessmnt"
## [1] 5465
## [1] "removing: faci_bun_t_audit"
## [1] 5465
## [1] "removing: faci_bun_t_basic"
## [1] 5465
## [1] "removing: faci_bun_t_behav"
## [1] 5465
## [1] "removing: faci_bun_t_bus"
## [1] 5465
## [1] "removing: faci_bun_t_care"
## [1] 5465
## [1] "removing: faci_bun_t_chng"
## [1] 5465
## [1] "removing: faci_bun_t_computer"
## [1] 5465
## [1] "removing: faci_bun_t_conf"
## [1] 5465
## [1] "removing: faci_bun_t_consultation"
## [1] 5465
## [1] "removing: faci_bun_t_counseling"
## [1] 5465
## [1] "removing: faci_bun_t_critical"
## [1] 5465
## [1] "removing: faci_bun_t_data"
## [1] 5465
## [1] "removing: faci_bun_t_direct"
## [1] 5465
## [1] "removing: faci_bun_t_disability"
## [1] 5465
## [1] "removing: faci_bun_t_discharge"
## [1] 5465
## [1] "removing: faci_bun_t_dommicl"
## [1] 5465
## [1] "removing: faci_bun_t_exam"
## [1] 5465
## [1] "removing: faci_bun_t_fac"
## [1] 5465
## [1] "removing: faci_bun_t_facility"
## [1] 5465
## [1] "removing: faci_bun_t_home"
## [1] 5464
## [1] "removing: faci_bun_t_hospice"
## [1] 5464
## [1] "removing: faci_bun_t_hospital"
## [1] 5464
## [1] "removing: faci_bun_t_initial"
## [1] 5464
## [1] "removing: faci_bun_t_inpatient"
## [1] 5464
## [1] "removing: faci_bun_t_interfacility"
## [1] 5464
## [1] "removing: faci_bun_t_interpof"
## [1] 5464
## [1] "removing: faci_bun_t_life"
## [1] 5464
## [1] "removing: faci_bun_t_nonemergency"
## [1] 5464
## [1] "removing: faci_bun_t_noner"
## [1] 5464
## [1] "removing: faci_bun_t_nursing"
## [1] 5464
## [1] "removing: faci_bun_t_observation"
## [1] 5464
## [1] "removing: faci_bun_t_on_call"
## [1] 5464
## [1] "removing: faci_bun_t_online"
## [1] 5464
## [1] "removing: faci_bun_t_phlebotomy"
## [1] 5464
## [1] "removing: faci_bun_t_phone"
## [1] 5464
## [1] "removing: faci_bun_t_physician"
## [1] 5464
## [1] "removing: faci_bun_t_preventine"
## [1] 5464
## [1] "removing: faci_bun_t_prolong"
## [1] 5463
## [1] "removing: faci_bun_t_service"
## [1] 5463
## [1] "removing: faci_bun_t_smoking"
## [1] 5463
## [1] "removing: faci_bun_t_standby"
## [1] 5463
## [1] "removing: faci_bun_t_subsequent"
## [1] 5463
## [1] "removing: faci_bun_t_support"
## [1] 5463
## [1] "removing: faci_bun_t_taxi"
## [1] 5463
## [1] "removing: faci_bun_t_team"
## [1] 5463
## [1] "removing: faci_bun_t_transort"
## [1] 5463
## [1] "removing: faci_bun_t_travel"
## [1] 5463
## [1] "removing: faci_bun_t_van"
## [1] 5463toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.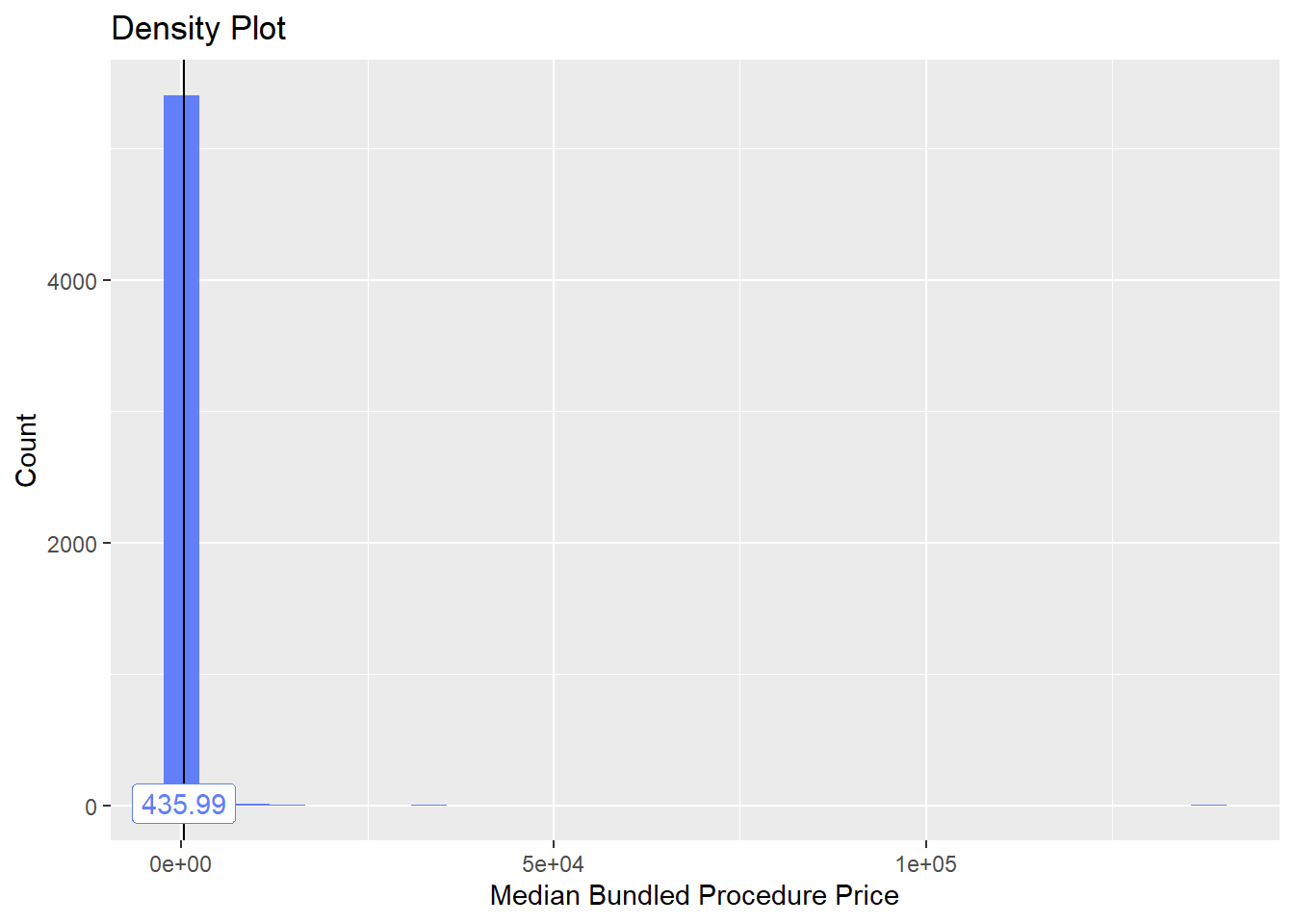
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 263 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_tum 0.534
## 2 surg_bun_t_foot 0.372
## 3 anes_bun_t_anes 0.327
## 4 anes_bun_t_anesth 0.322
## 5 surg_bun_t_toe 0.291
## 6 surg_bun_t_exc 0.285
## 7 anes_bun_t_surg 0.218
## 8 anes_bun_t_lower 0.217
## 9 surg_bun_t_compression 0.216
## 10 surg_bun_t_decompression 0.216
## # ... with 253 more rowstoe_nail2_clean %>% get_tag_density_information("surg_bun_t_tum") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_tum"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 9620## $dist_plots
##
## $stat_tables
toe_nail2_clean <- toe_nail2_clean[surg_bun_t_tum==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.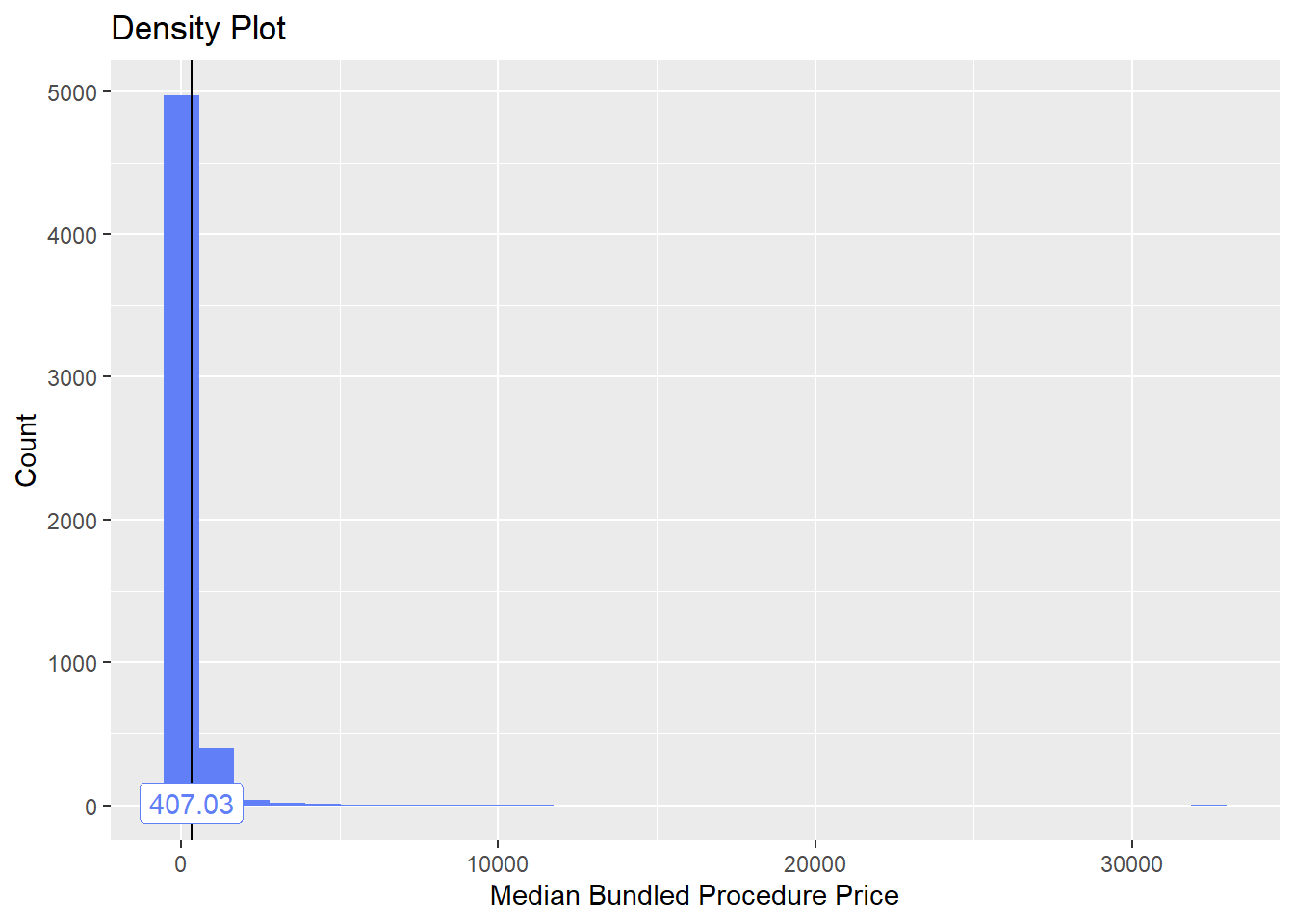
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 262 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_anes 0.606
## 2 surg_bun_t_compression 0.603
## 3 surg_bun_t_decompression 0.603
## 4 surg_bun_t_lower 0.603
## 5 surg_bun_t_release 0.603
## 6 anes_bun_t_surg 0.598
## 7 anes_bun_t_lower 0.594
## 8 anes_bun_t_anesth 0.588
## 9 medi_bun_t_choline 0.561
## 10 anes_bun_t_leg 0.544
## # ... with 252 more rowstoe_nail2_clean %>% get_tag_density_information("anes_bun_t_anes") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ anes_bun_t_anes"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 450## $dist_plots
##
## $stat_tables
toe_nail2_clean <- toe_nail2_clean[anes_bun_t_anes==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.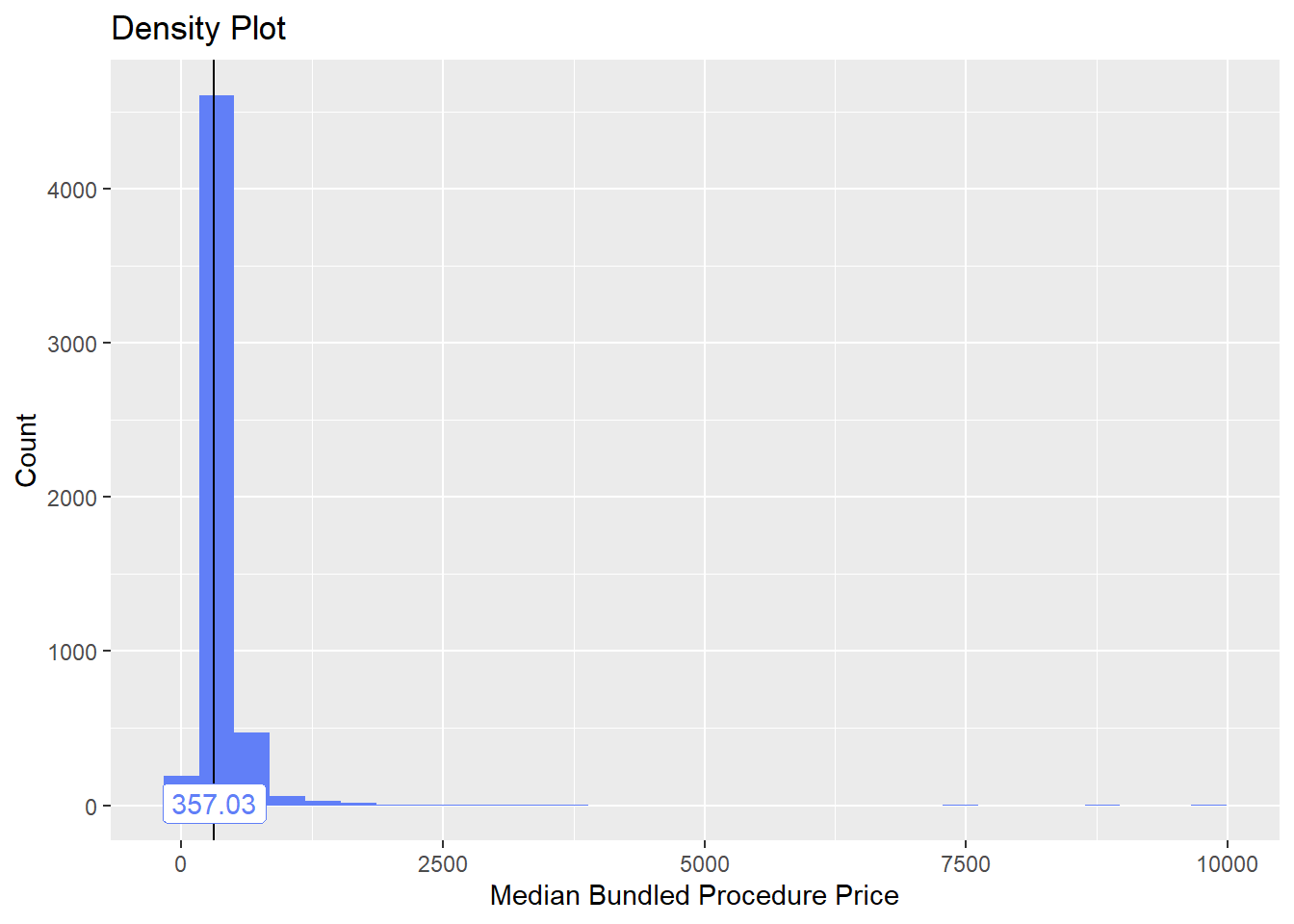
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 229 x 2
## name correlation
## <chr> <dbl>
## 1 medi_bun_t_wound 0.308
## 2 surg_bun_t_sub 0.271
## 3 medi_bun_t_analy 0.238
## 4 faci_bun_t_visit 0.229
## 5 faci_bun_t_dept 0.214
## 6 faci_bun_t_emergency 0.214
## 7 surg_bun_t_tiss 0.210
## 8 path_bun_t_immuno 0.209
## 9 faci_bun_t_pat 0.197
## 10 faci_bun_t_office 0.197
## # ... with 219 more rowstoe_nail2_clean %>% get_tag_density_information("medi_bun_t_wound") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_wound"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 517## $dist_plots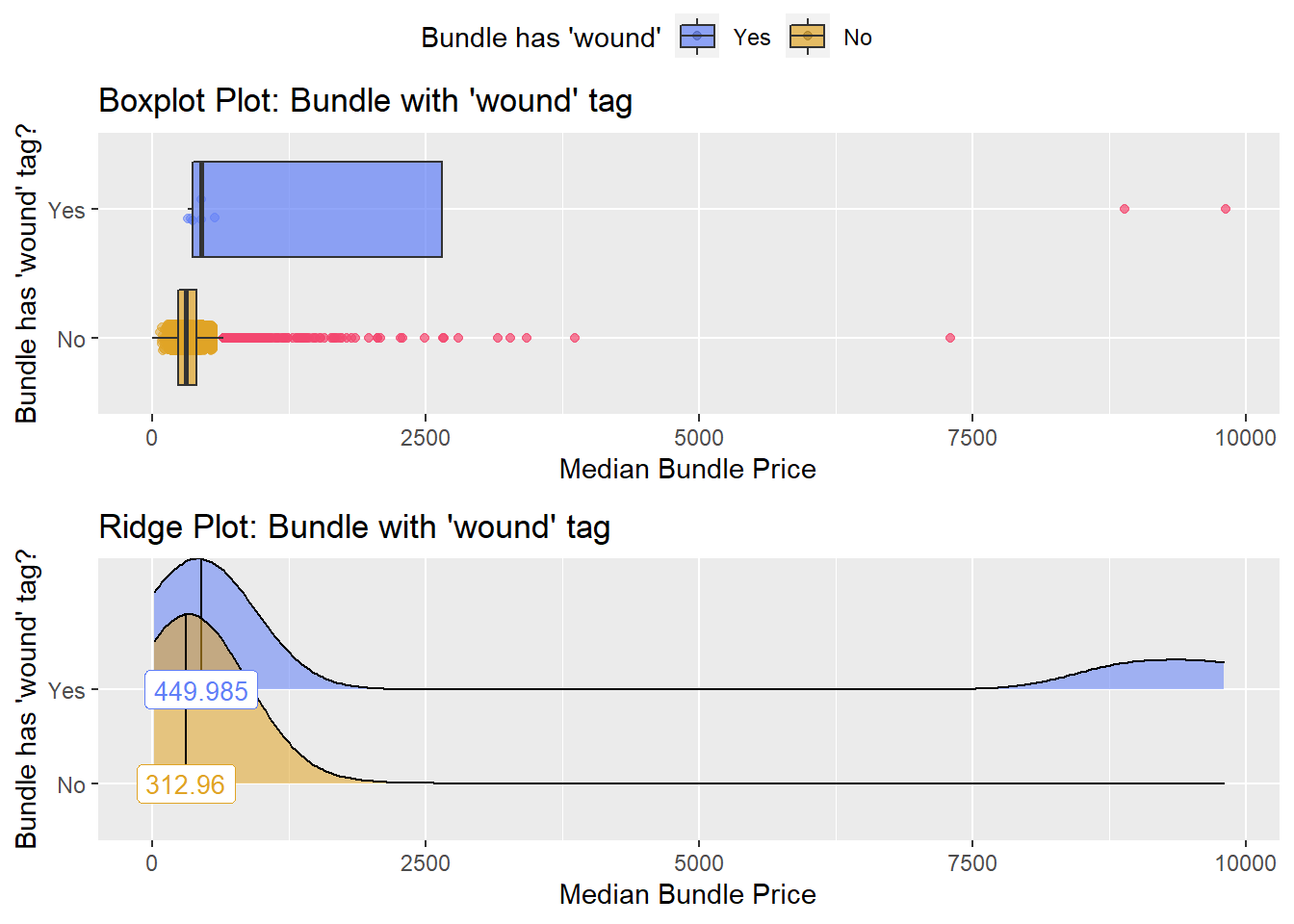
##
## $stat_tables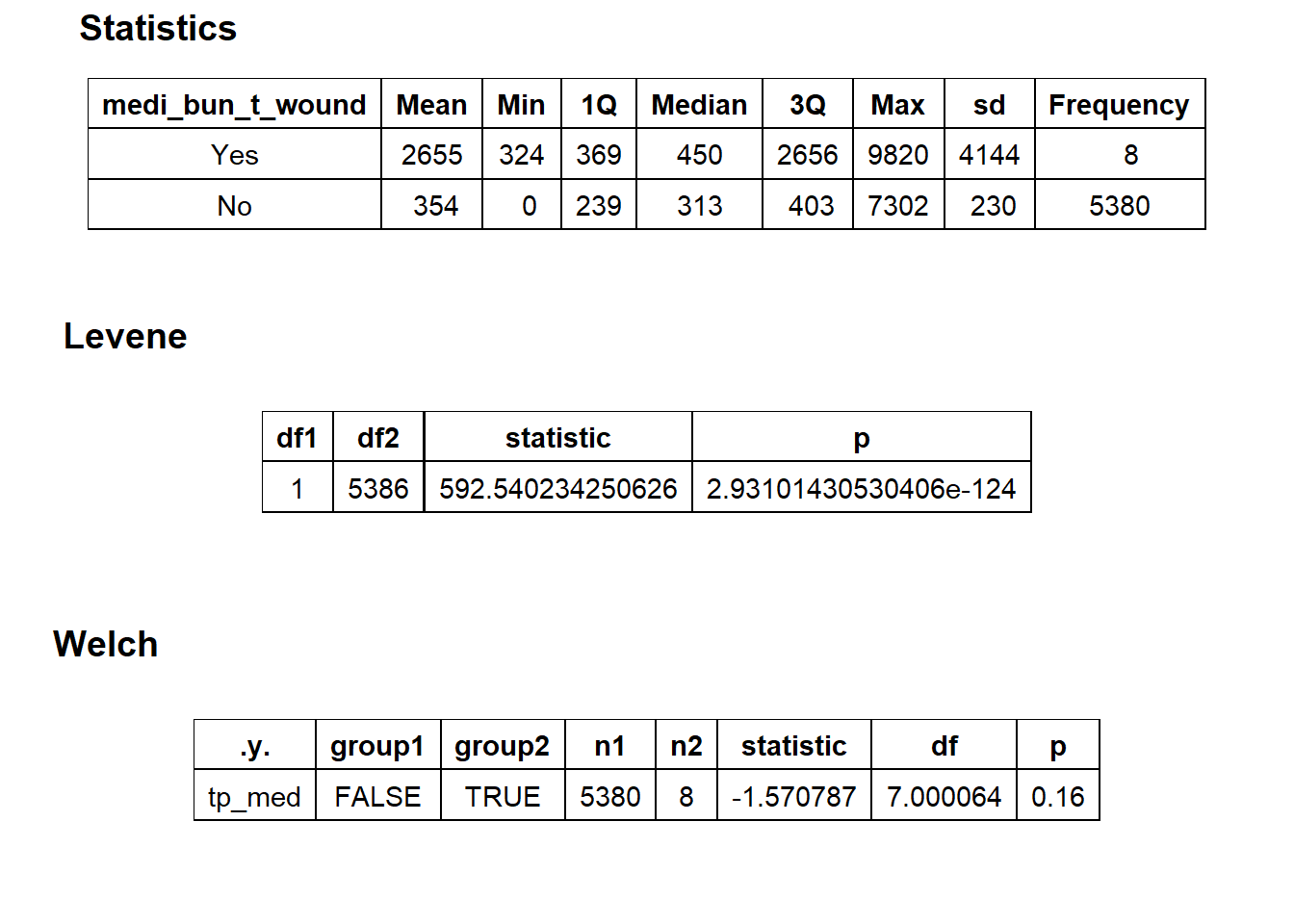
toe_nail2_clean <- toe_nail2_clean[medi_bun_t_wound==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.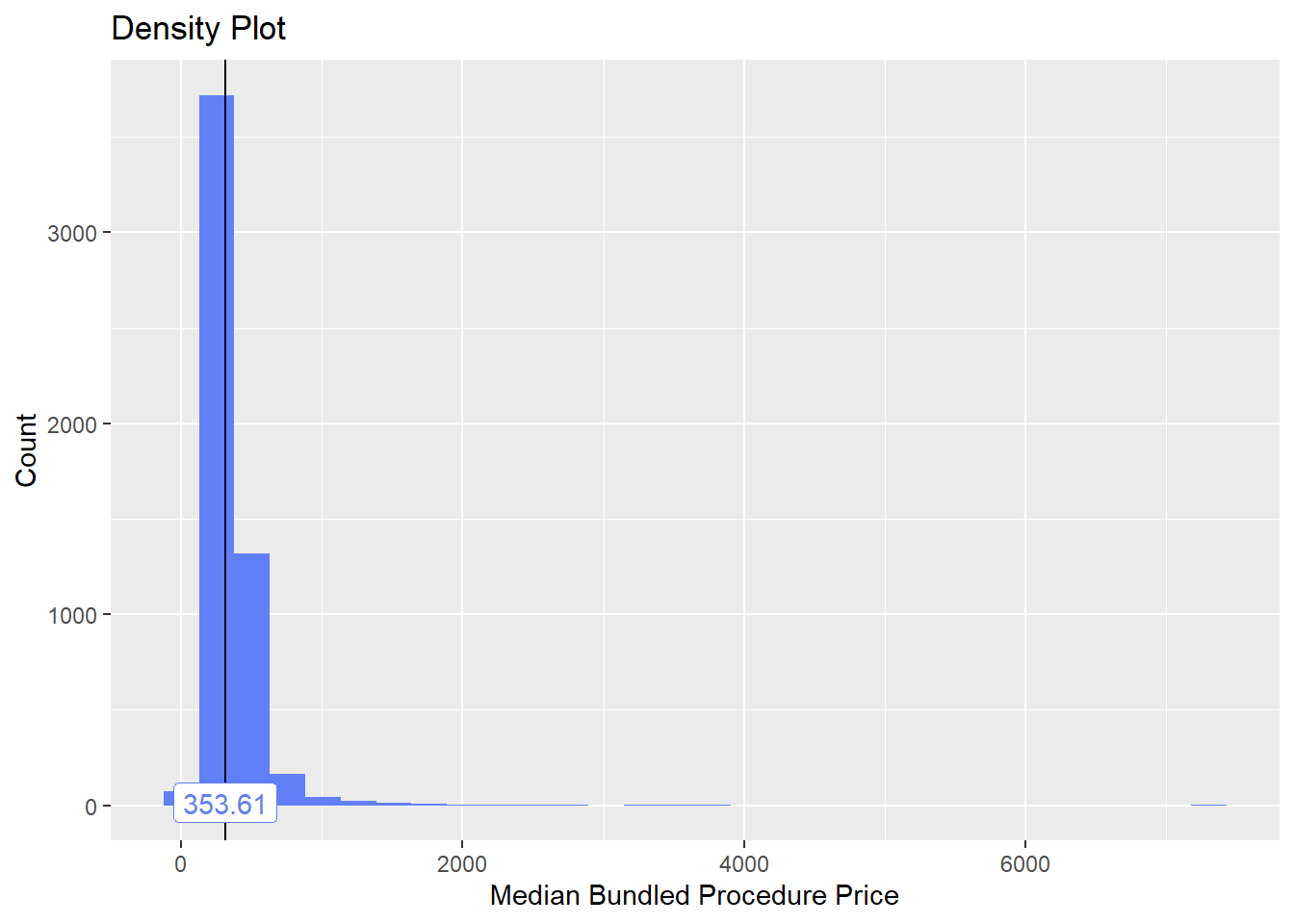
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 228 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.275
## 2 faci_bun_t_dept 0.270
## 3 faci_bun_t_emergency 0.270
## 4 path_bun_t_patho 0.246
## 5 path_bun_t_pathologist 0.246
## 6 path_bun_t_tissue 0.239
## 7 faci_bun_t_pat 0.235
## 8 faci_bun_t_office 0.235
## 9 path_bun_t_detect 0.221
## 10 path_bun_t_agent 0.219
## # ... with 218 more rowstoe_nail2_clean %>% get_tag_density_information("faci_bun_t_emergency") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ faci_bun_t_emergency"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 157## $dist_plots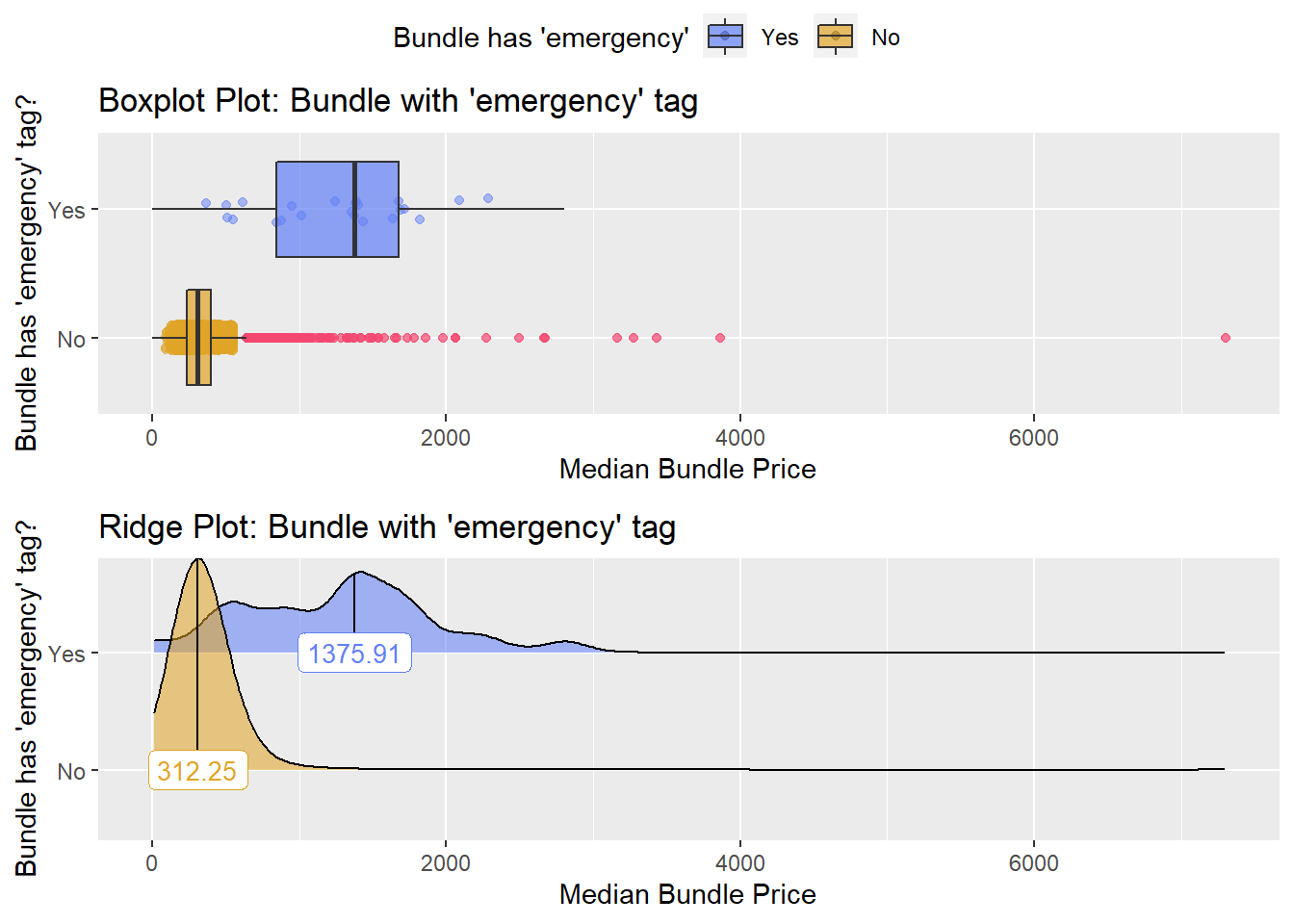
##
## $stat_tables
toe_nail2_clean <- toe_nail2_clean[faci_bun_t_emergency==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.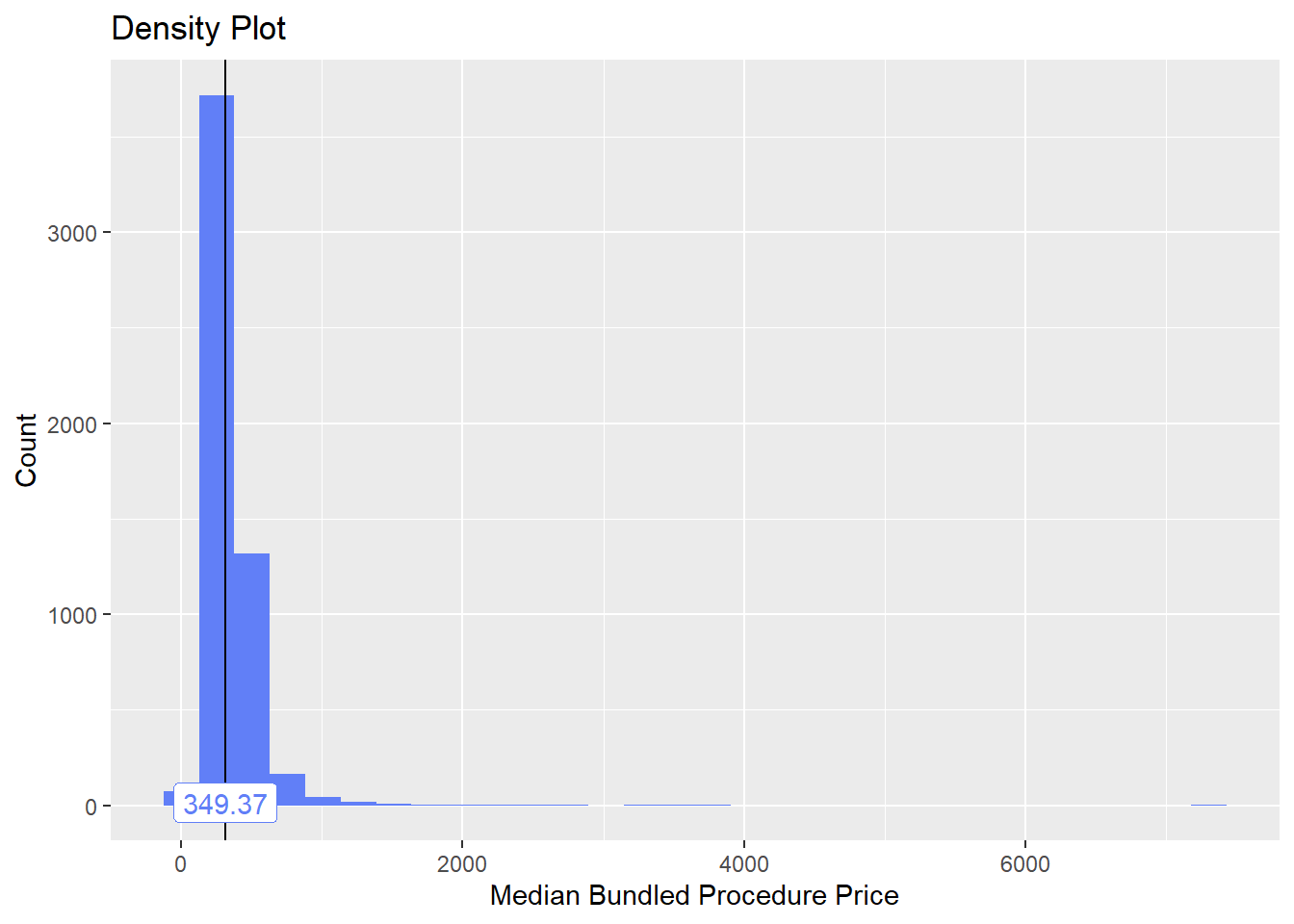
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 219 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.276
## 2 faci_bun_t_pat 0.265
## 3 faci_bun_t_office 0.265
## 4 path_bun_t_patho 0.263
## 5 path_bun_t_pathologist 0.263
## 6 path_bun_t_tissue 0.255
## 7 path_bun_t_detect 0.235
## 8 path_bun_t_agent 0.233
## 9 path_bun_t_dna 0.222
## 10 path_bun_t_drug 0.157
## # ... with 209 more rowstoe_nail2_clean %>% get_tag_density_information("path_bun_t_patho") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_patho"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 104## $dist_plots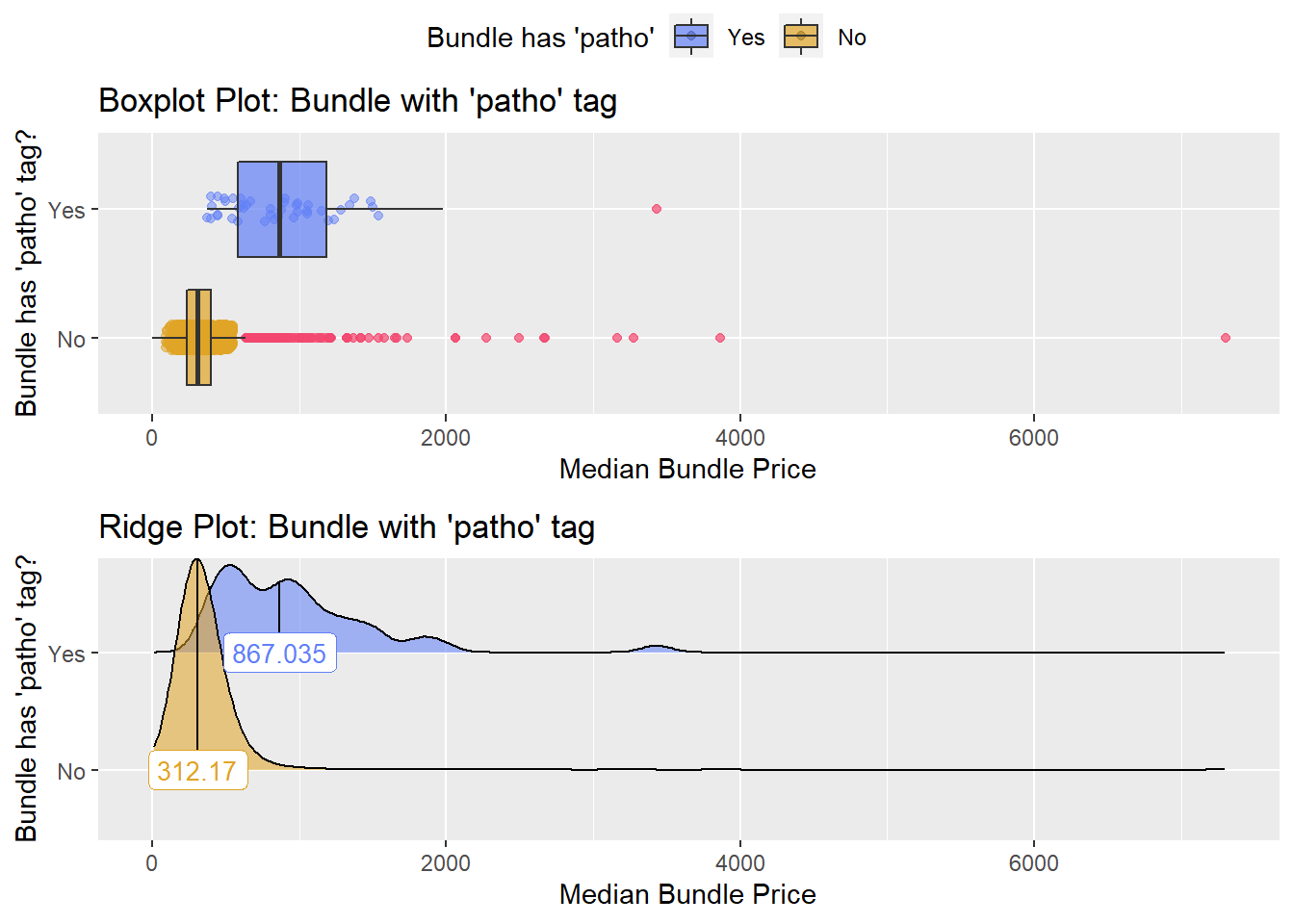
##
## $stat_tables
toe_nail2_clean <- toe_nail2_clean[path_bun_t_patho==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.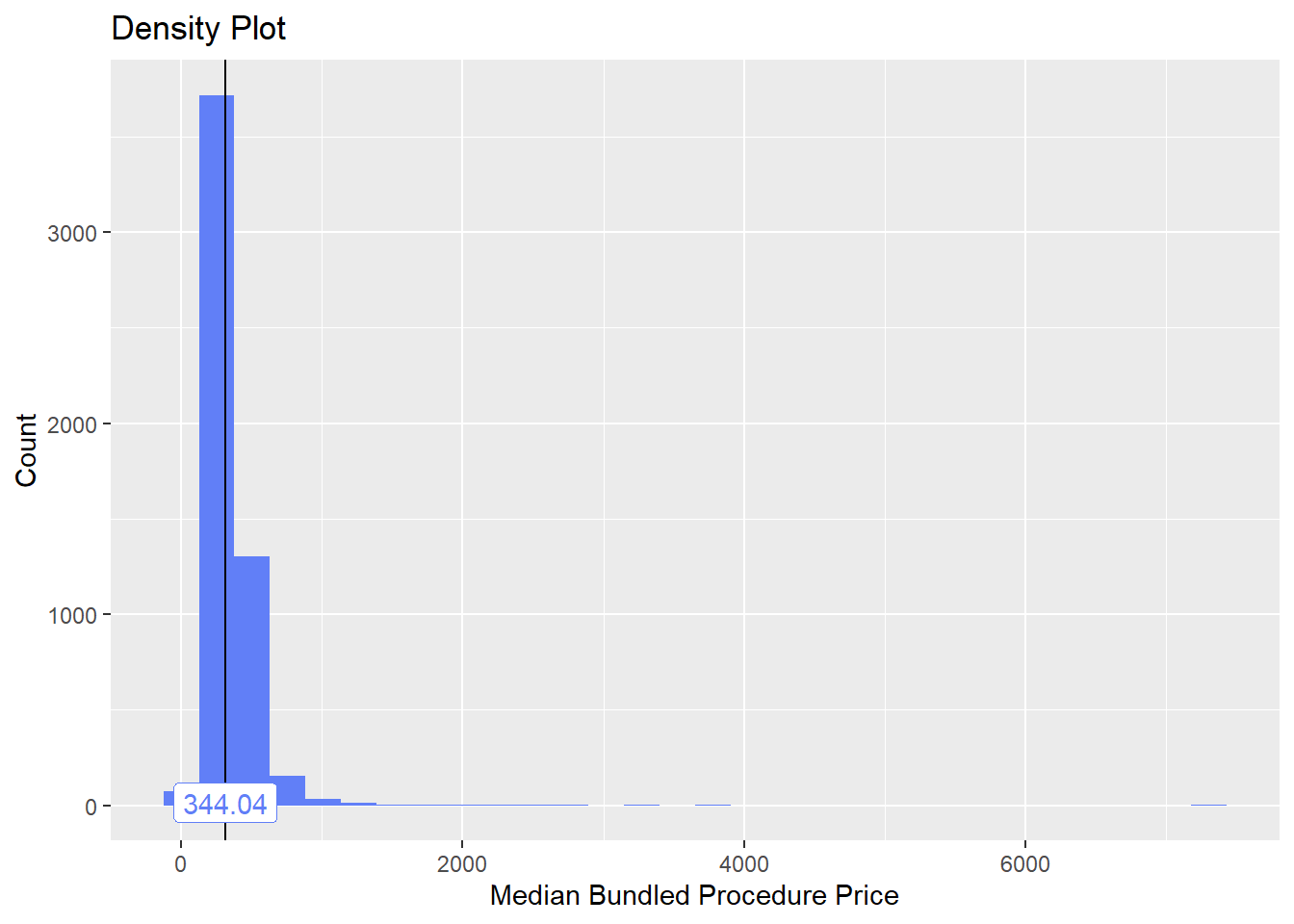
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 216 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.284
## 2 faci_bun_t_pat 0.272
## 3 faci_bun_t_office 0.272
## 4 path_bun_t_drug 0.168
## 5 surg_bun_t_debrid 0.165
## 6 surg_bun_t_debride 0.165
## 7 path_bun_t_test 0.133
## 8 path_bun_t_lipid 0.131
## 9 medi_bun_t_inject 0.127
## 10 faci_bun_t_est 0.119
## # ... with 206 more rowstoe_nail2_clean %>% get_tag_density_information("path_bun_t_drug") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_drug"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 522## $dist_plots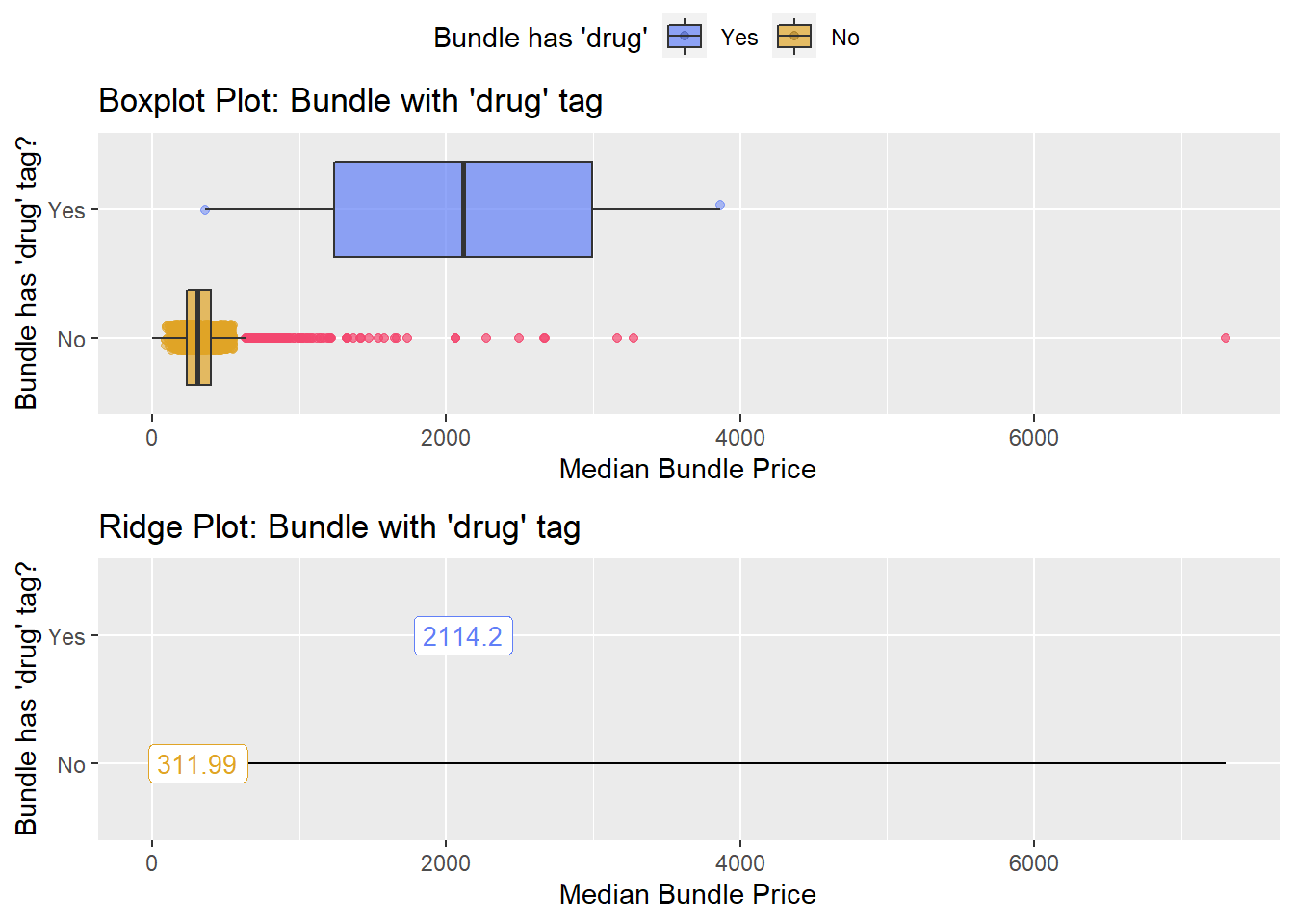
##
## $stat_tables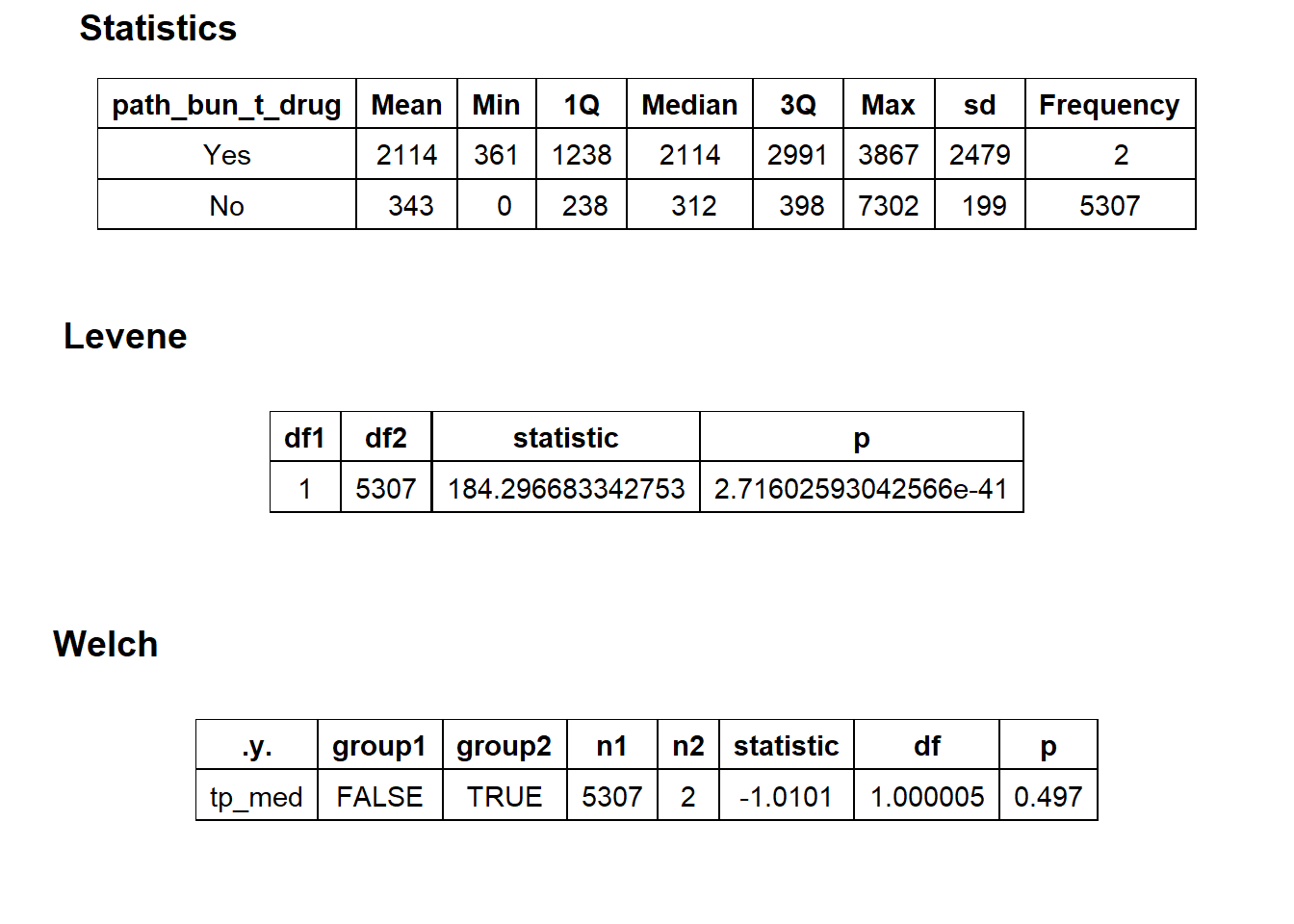
toe_nail2_clean <- toe_nail2_clean[path_bun_t_drug==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 215 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.290
## 2 faci_bun_t_pat 0.278
## 3 faci_bun_t_office 0.277
## 4 surg_bun_t_debrid 0.170
## 5 surg_bun_t_debride 0.170
## 6 path_bun_t_lipid 0.135
## 7 medi_bun_t_inject 0.131
## 8 radi_bun_t_exam 0.123
## 9 medi_bun_t_stud 0.120
## 10 path_bun_t_panel 0.119
## # ... with 205 more rowstoe_nail2_clean %>% get_tag_density_information("surg_bun_t_debrid") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_debrid"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 63.3## $dist_plots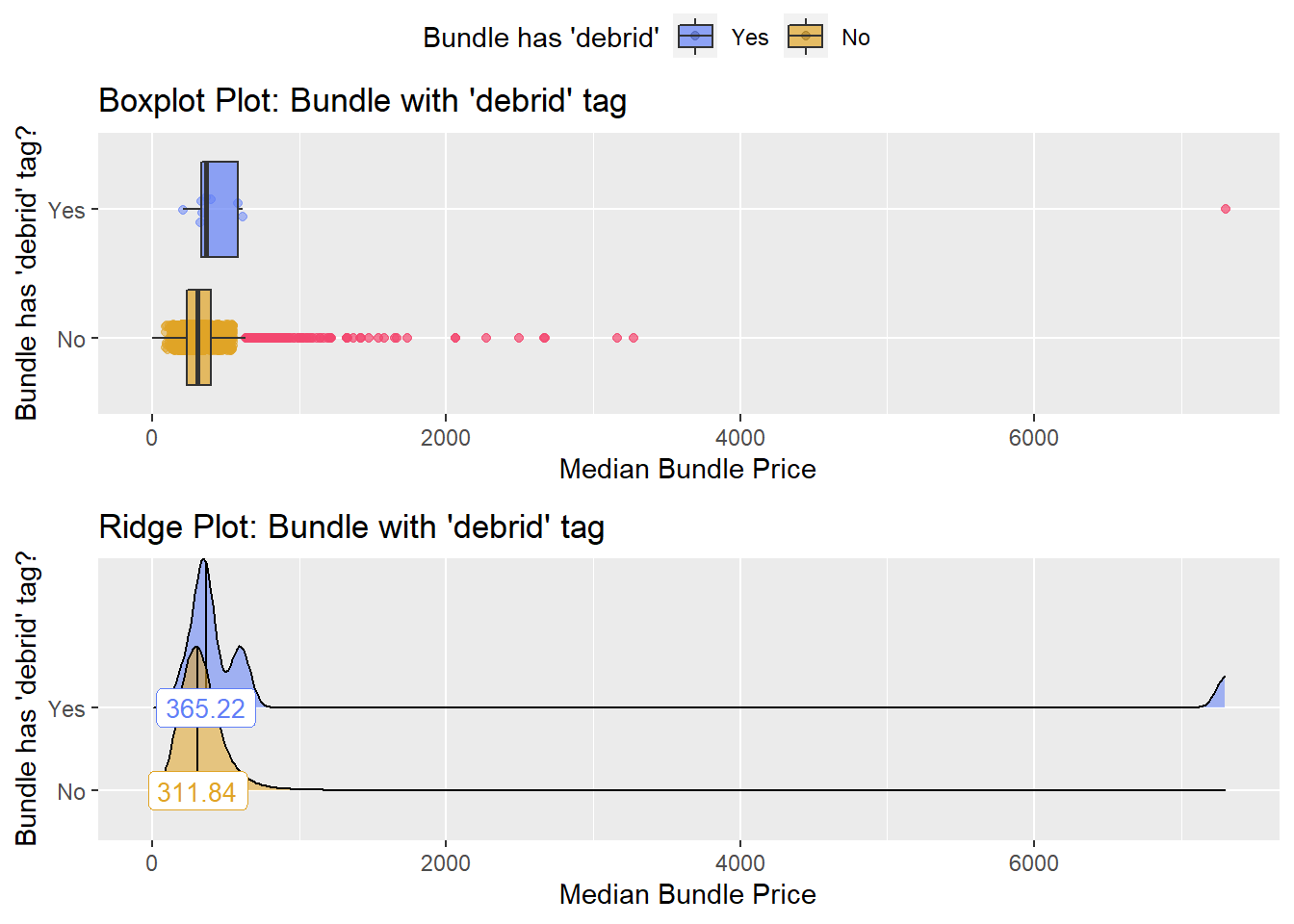
##
## $stat_tables
toe_nail2_clean <- toe_nail2_clean[surg_bun_t_debrid==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.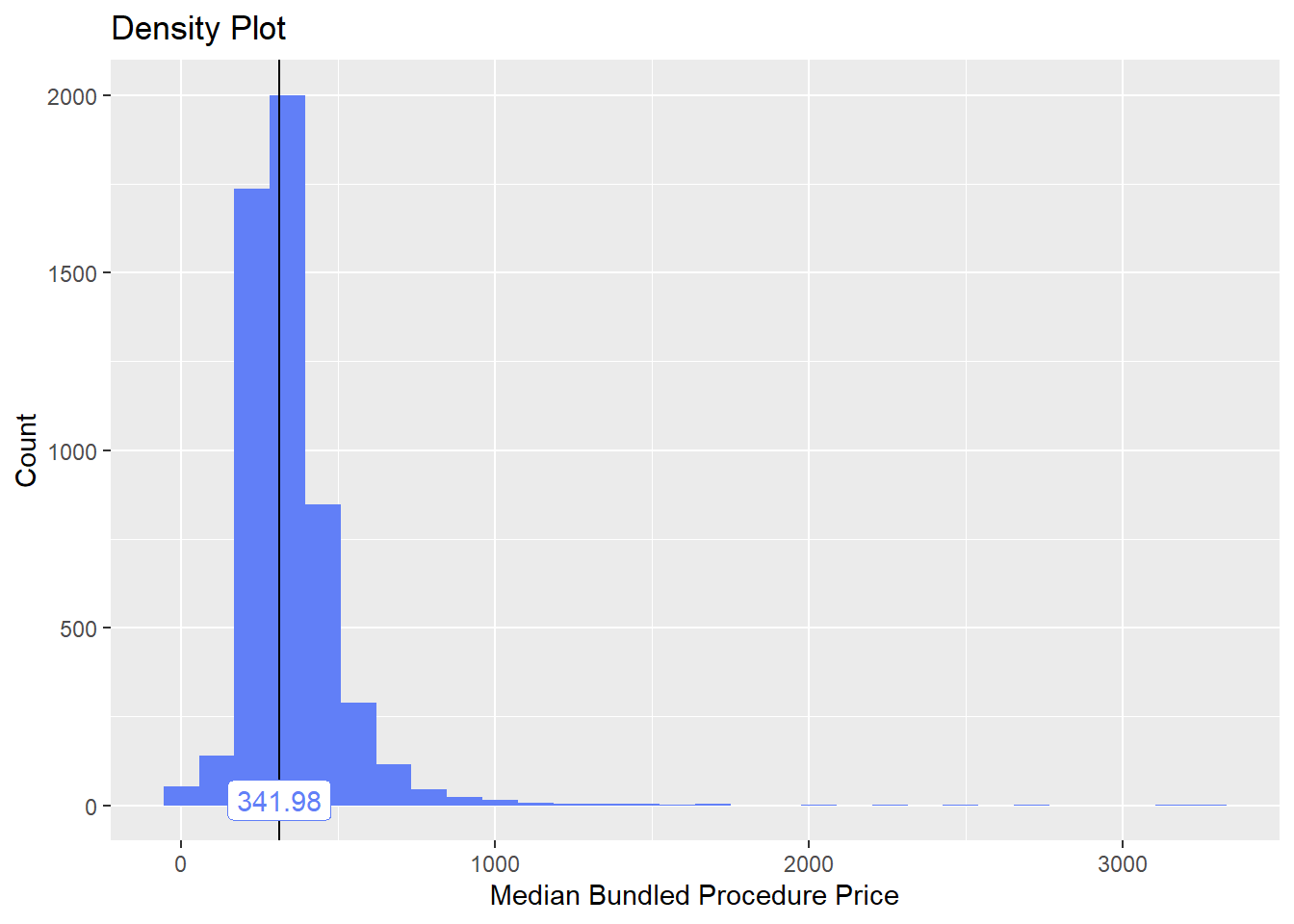
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 213 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.34
## 2 faci_bun_t_pat 0.326
## 3 faci_bun_t_office 0.325
## 4 path_bun_t_lipid 0.155
## 5 radi_bun_t_exam 0.140
## 6 medi_bun_t_stud 0.137
## 7 faci_bun_t_est 0.137
## 8 path_bun_t_panel 0.137
## 9 radi_bun_t_x-ray 0.136
## 10 surg_bun_t_sub 0.135
## # ... with 203 more rowstoe_nail2_clean %>% get_tag_density_information("path_bun_t_lipid") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_lipid"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 73.7## $dist_plots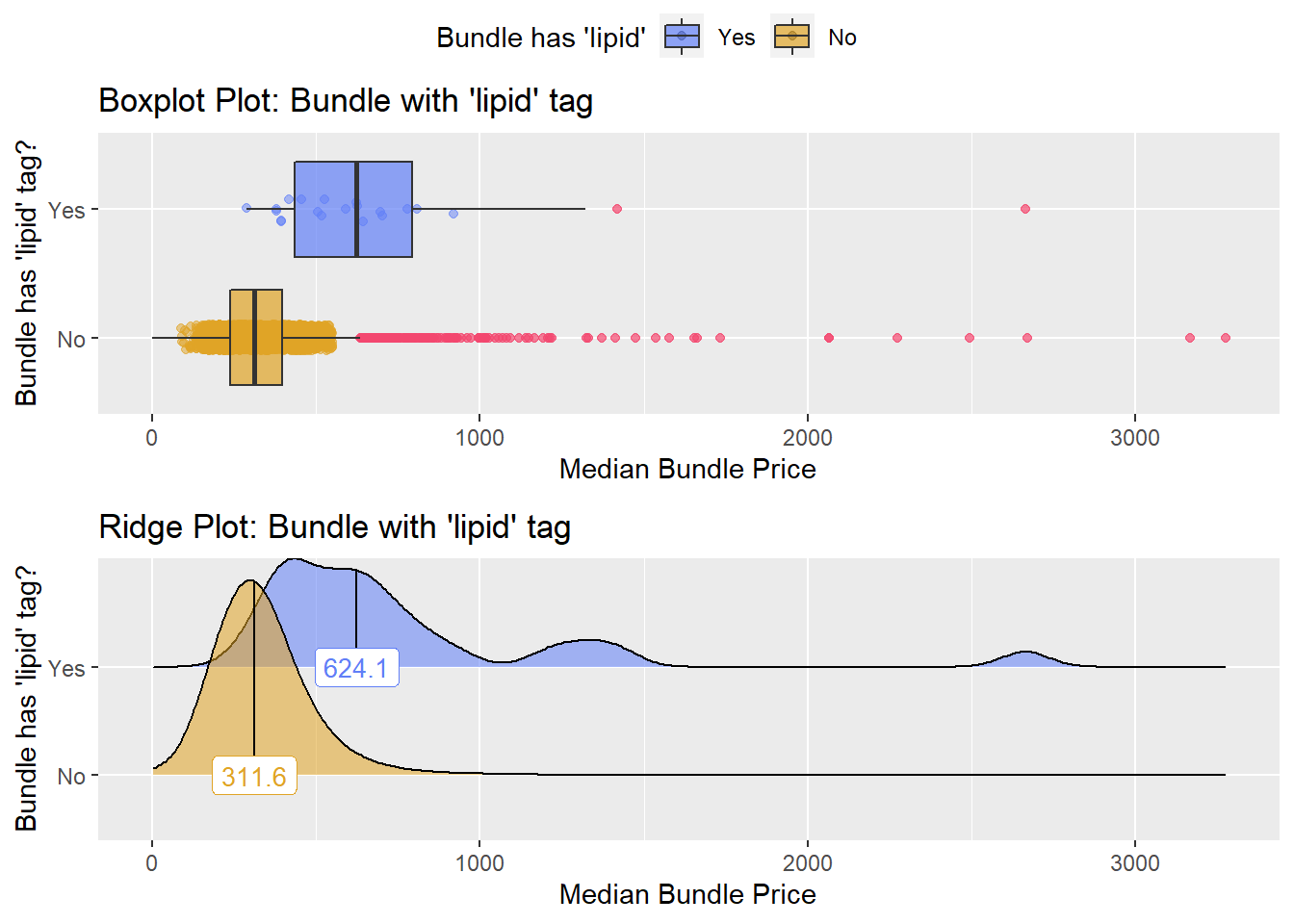
##
## $stat_tables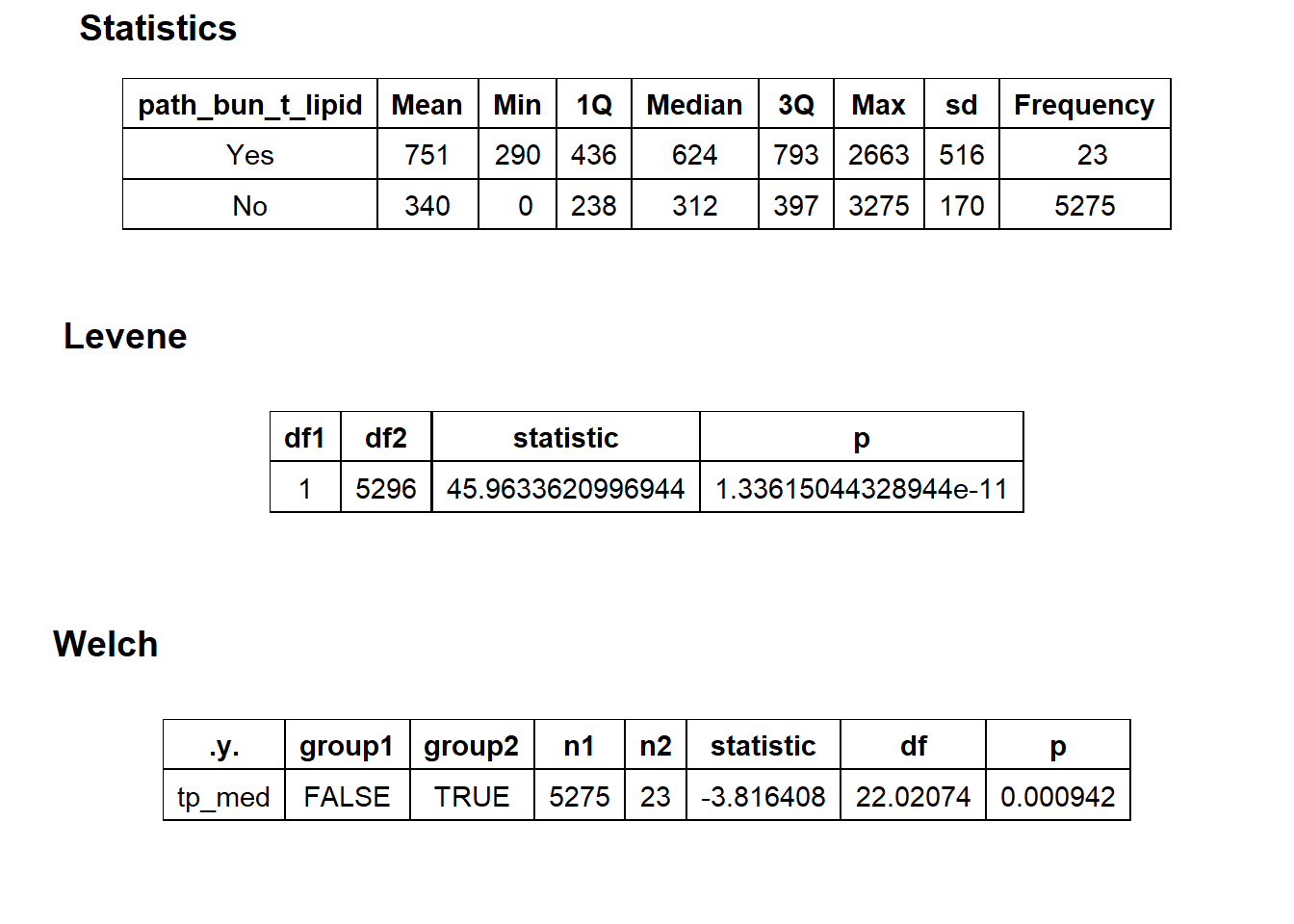
toe_nail2_clean <- toe_nail2_clean[path_bun_t_lipid==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 211 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.343
## 2 faci_bun_t_pat 0.337
## 3 faci_bun_t_office 0.337
## 4 medi_bun_t_stud 0.142
## 5 surg_bun_t_sub 0.140
## 6 radi_bun_t_exam 0.135
## 7 radi_bun_t_x-ray 0.132
## 8 faci_bun_t_est 0.128
## 9 radi_bun_t_lumbar 0.12
## 10 radi_bun_t_without_dye 0.119
## # ... with 201 more rowstoe_nail2_clean %>% get_tag_density_information("medi_bun_t_stud") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_stud"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 174## $dist_plots
##
## $stat_tables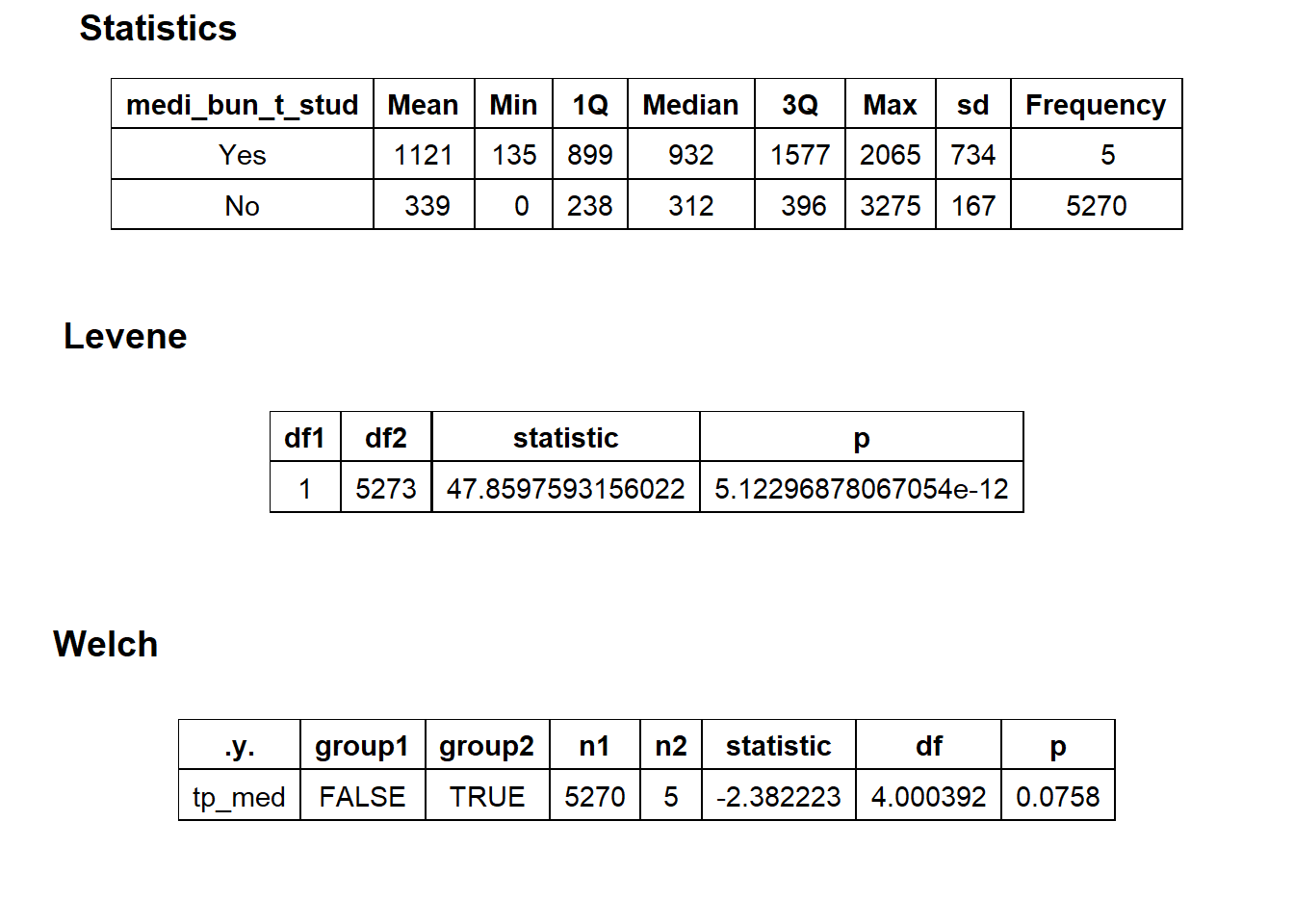
toe_nail2_clean <- toe_nail2_clean[medi_bun_t_stud==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.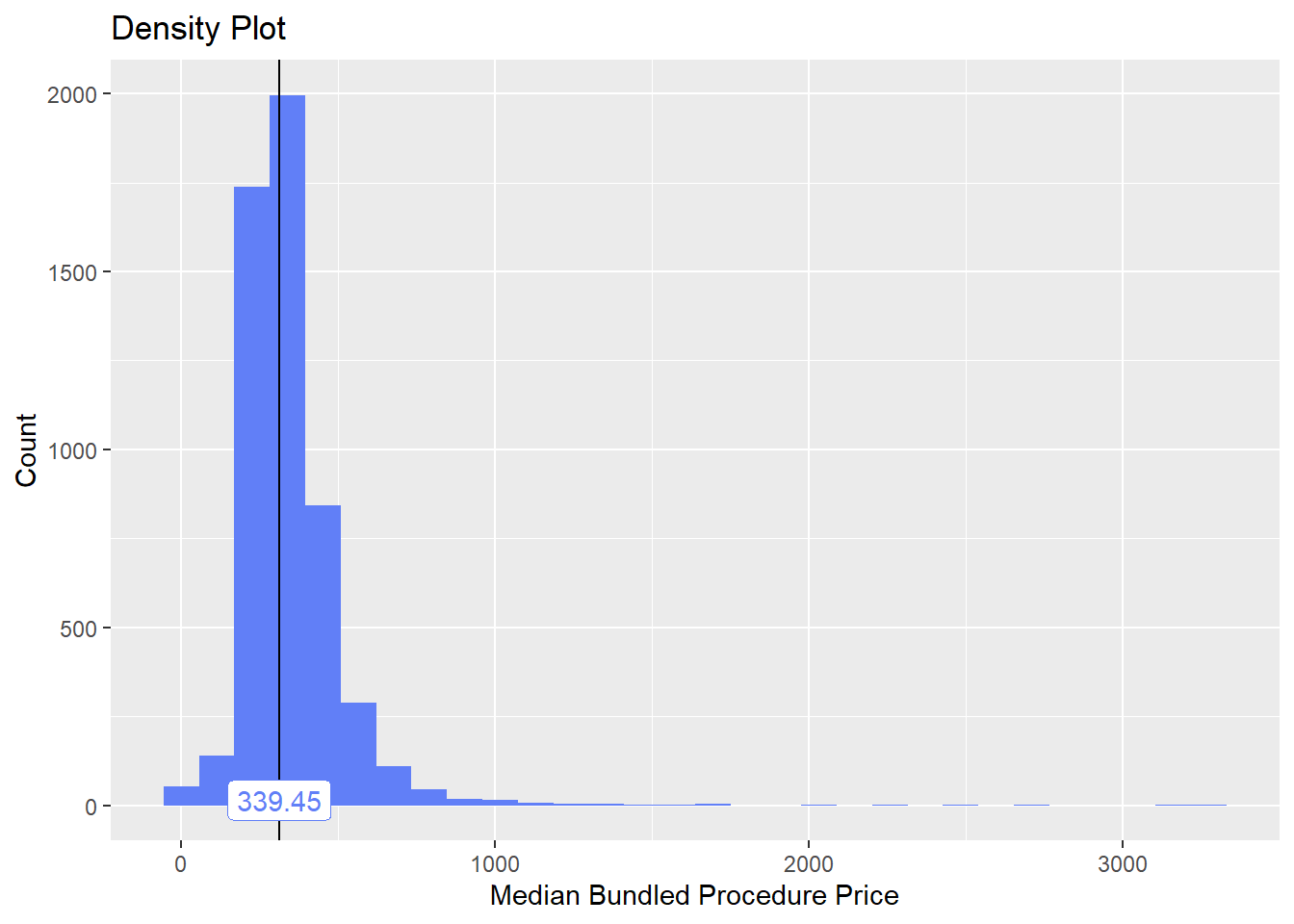
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 210 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.347
## 2 faci_bun_t_pat 0.340
## 3 faci_bun_t_office 0.340
## 4 surg_bun_t_sub 0.142
## 5 radi_bun_t_exam 0.138
## 6 radi_bun_t_x-ray 0.135
## 7 faci_bun_t_est 0.124
## 8 radi_bun_t_lumbar 0.122
## 9 radi_bun_t_without_dye 0.122
## 10 path_bun_t_detect 0.118
## # ... with 200 more rowstoe_nail2_clean %>% get_tag_density_information("surg_bun_t_sub") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_sub"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 411## $dist_plots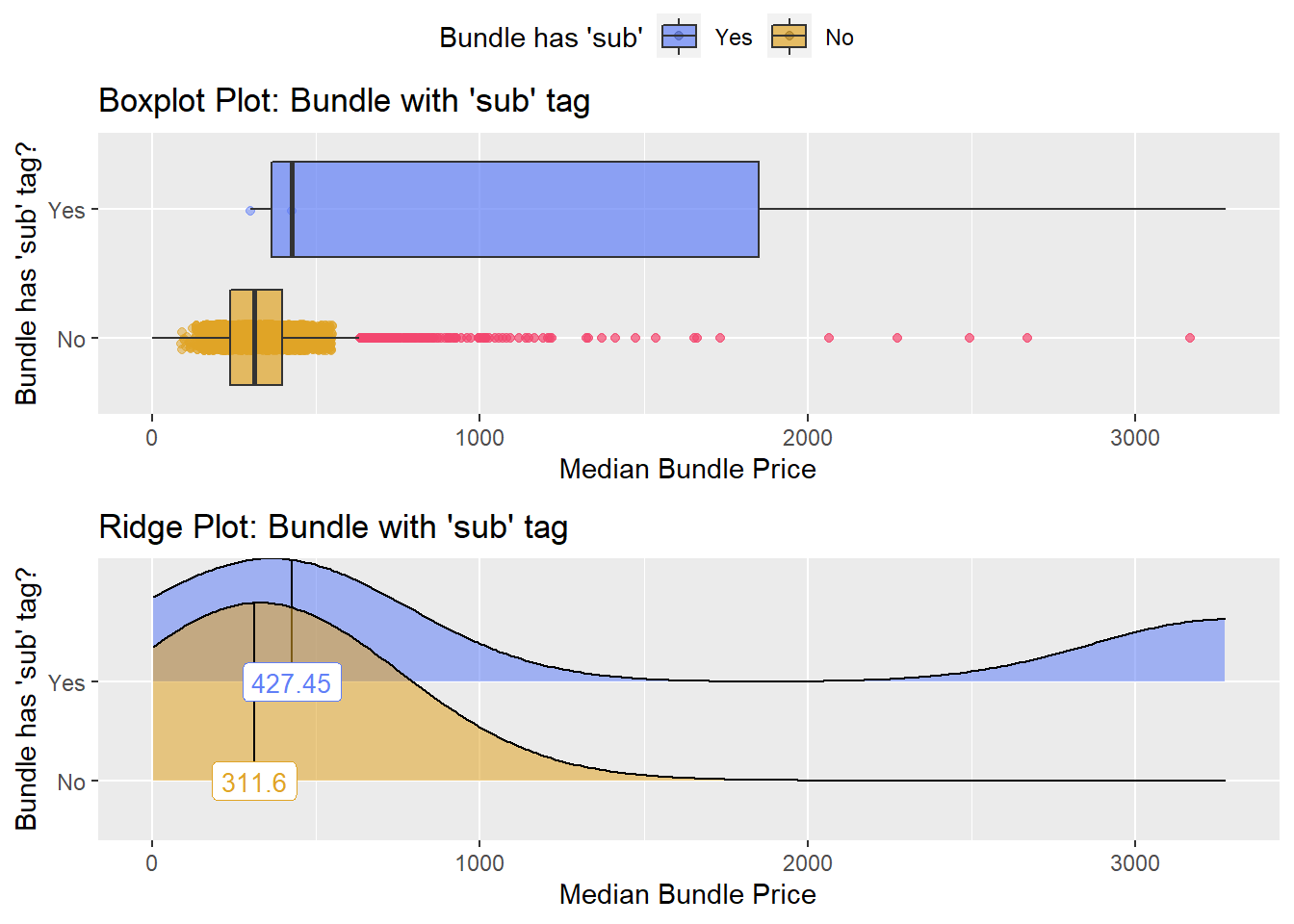
##
## $stat_tables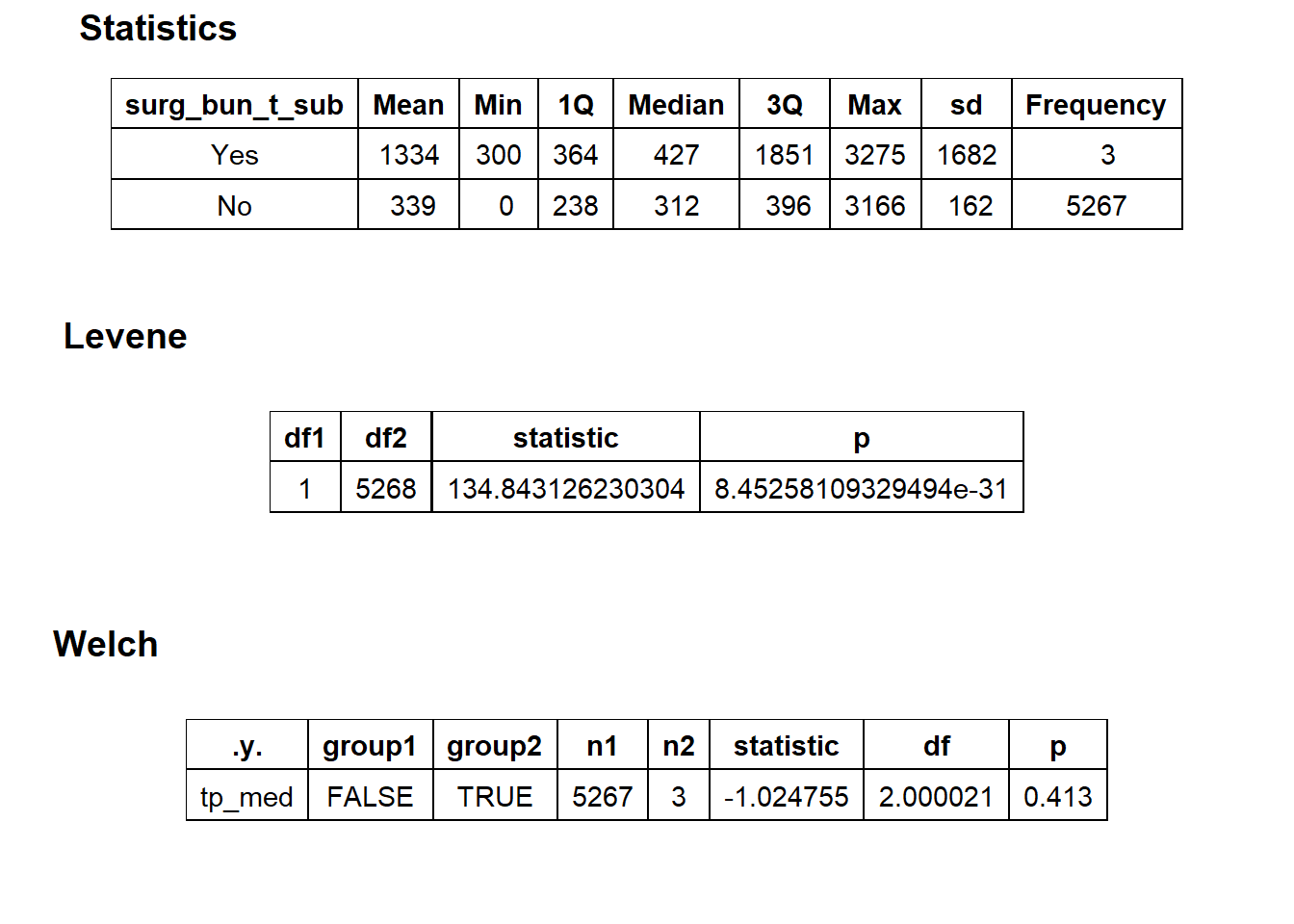
toe_nail2_clean <- toe_nail2_clean[surg_bun_t_sub==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.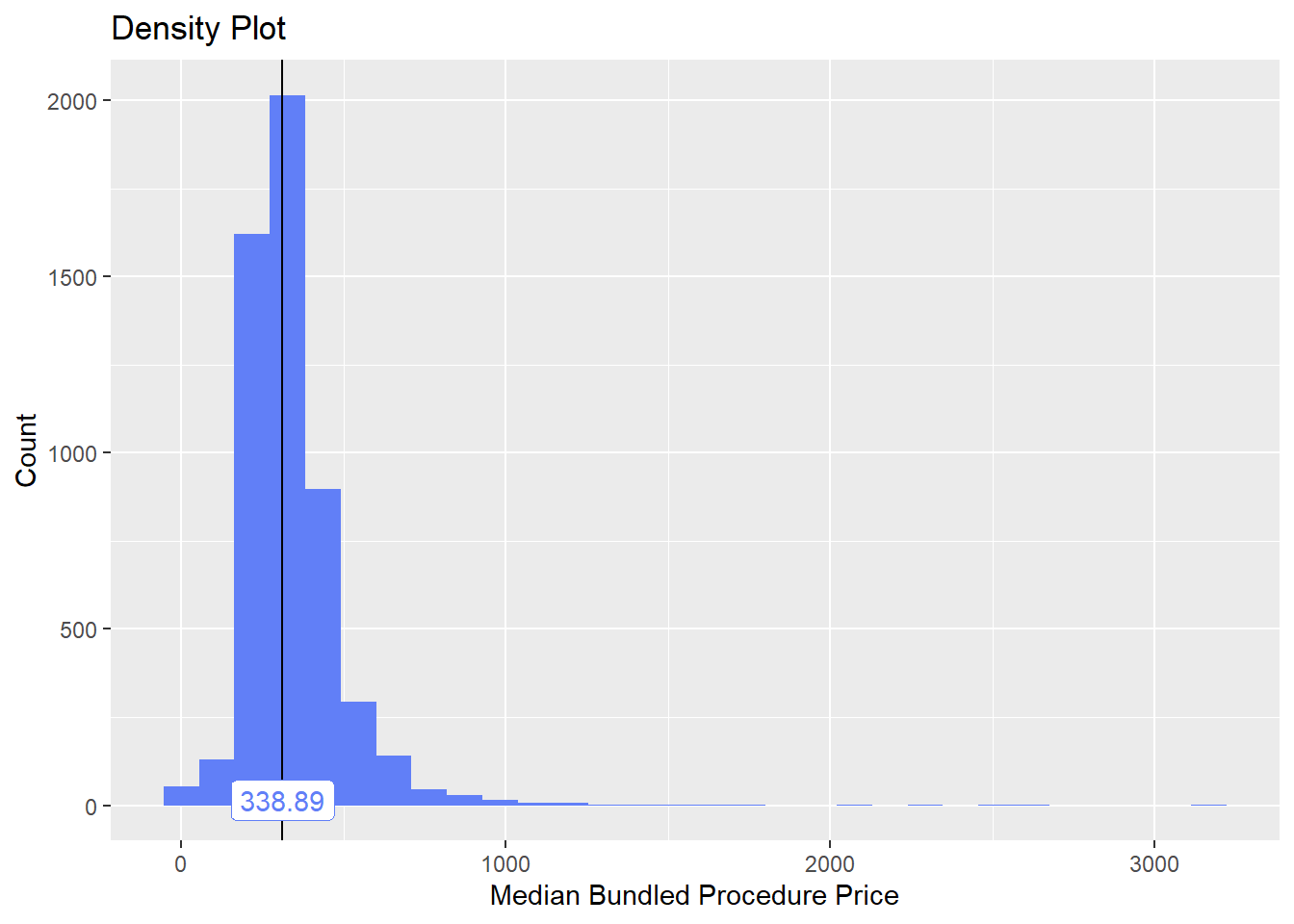
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 209 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.355
## 2 faci_bun_t_pat 0.348
## 3 faci_bun_t_office 0.348
## 4 radi_bun_t_exam 0.143
## 5 radi_bun_t_x-ray 0.140
## 6 radi_bun_t_lumbar 0.126
## 7 radi_bun_t_without_dye 0.126
## 8 path_bun_t_detect 0.122
## 9 radi_bun_t_mri 0.122
## 10 faci_bun_t_est 0.122
## # ... with 199 more rowstoe_nail2_clean %>% get_tag_density_information("radi_bun_t_lumbar") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ radi_bun_t_lumbar"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 270## $dist_plots
##
## $stat_tables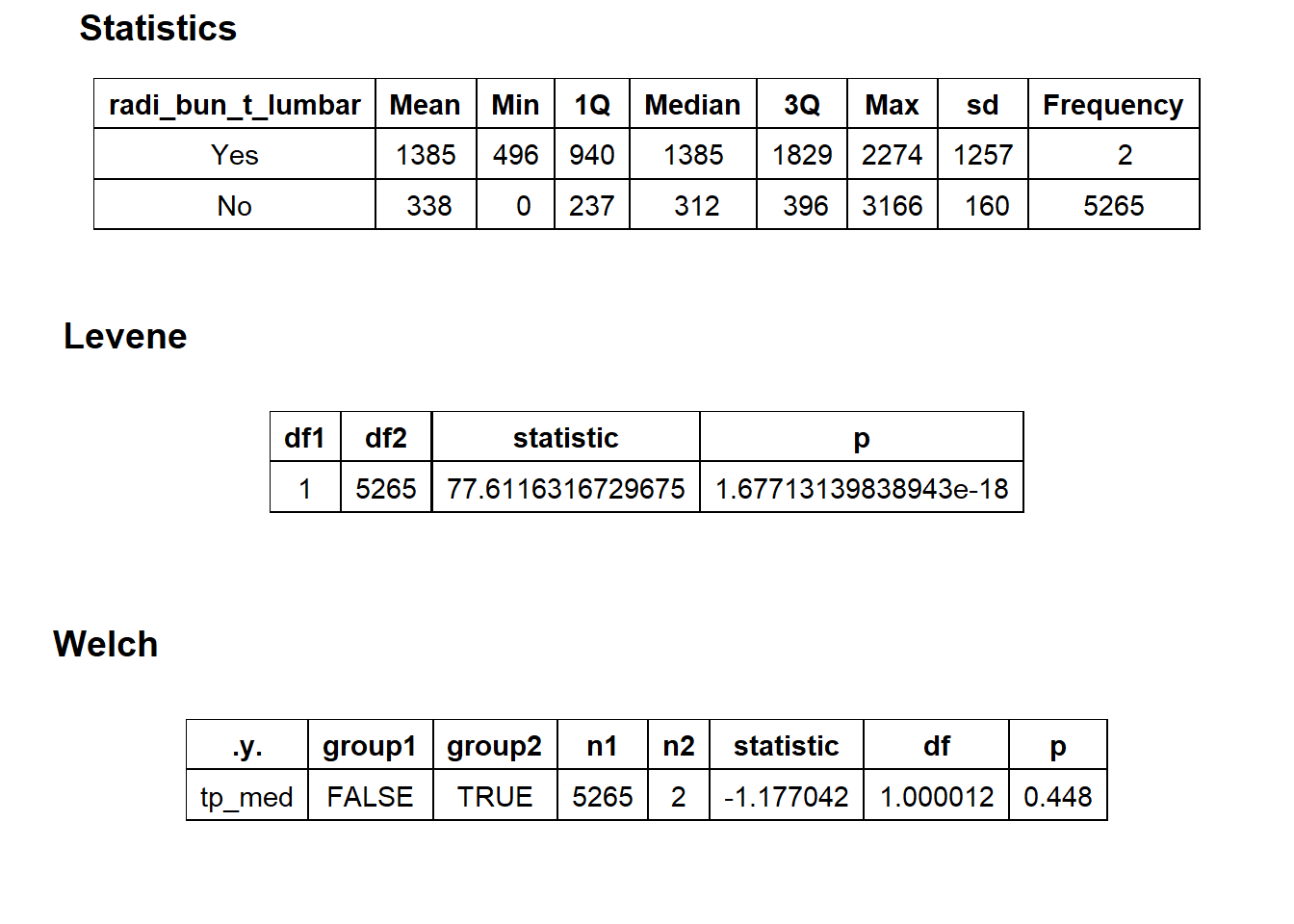
toe_nail2_clean <- toe_nail2_clean[radi_bun_t_lumbar==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.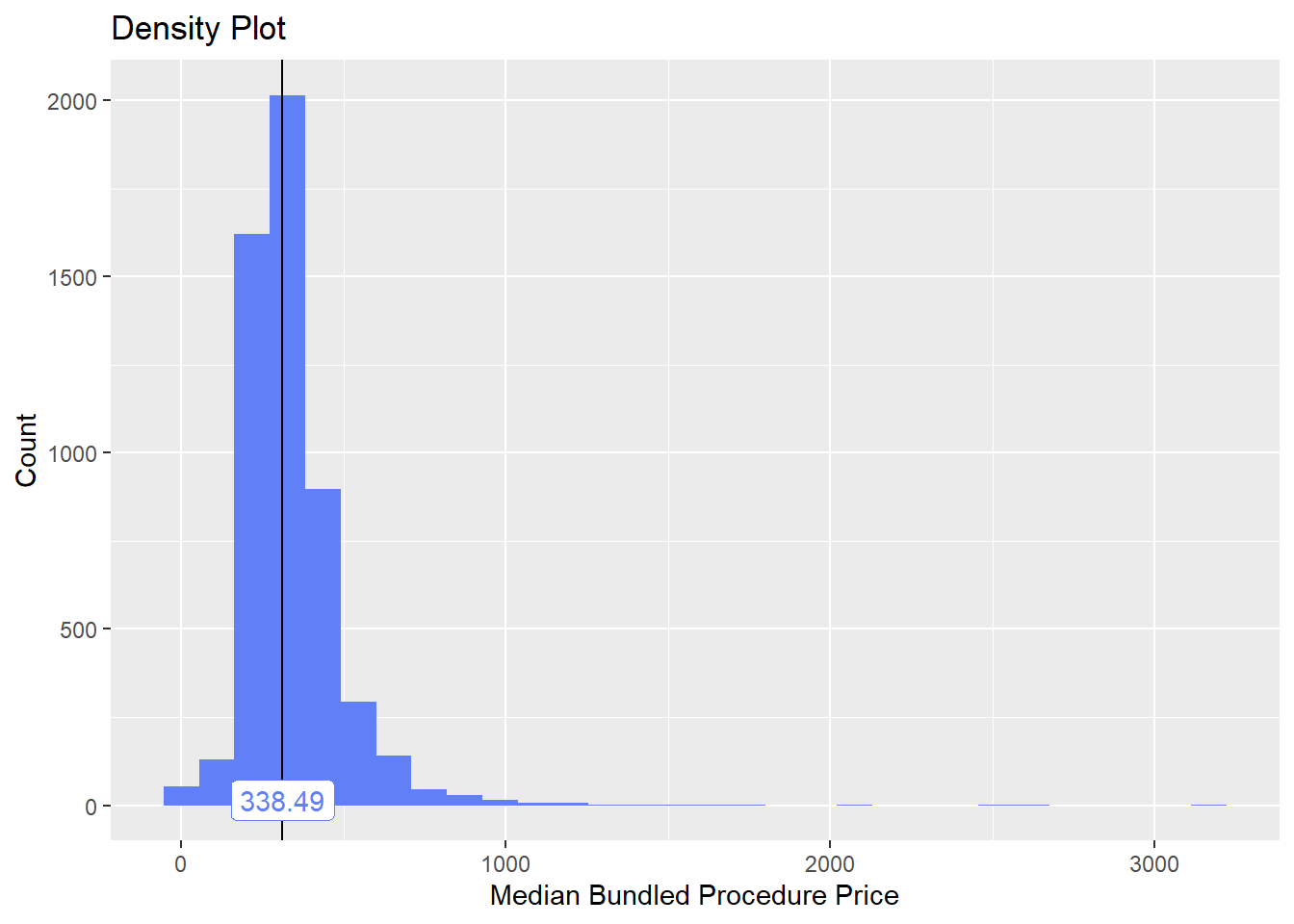
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 208 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.363
## 2 faci_bun_t_pat 0.356
## 3 faci_bun_t_office 0.356
## 4 radi_bun_t_exam 0.145
## 5 radi_bun_t_x-ray 0.142
## 6 faci_bun_t_est 0.125
## 7 path_bun_t_detect 0.124
## 8 path_bun_t_agent 0.122
## 9 path_bun_t_dna 0.121
## 10 radi_bun_t_foot 0.109
## # ... with 198 more rowstoe_nail2_clean %>% get_tag_density_information("radi_bun_t_exam") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ radi_bun_t_exam"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 30.7## $dist_plots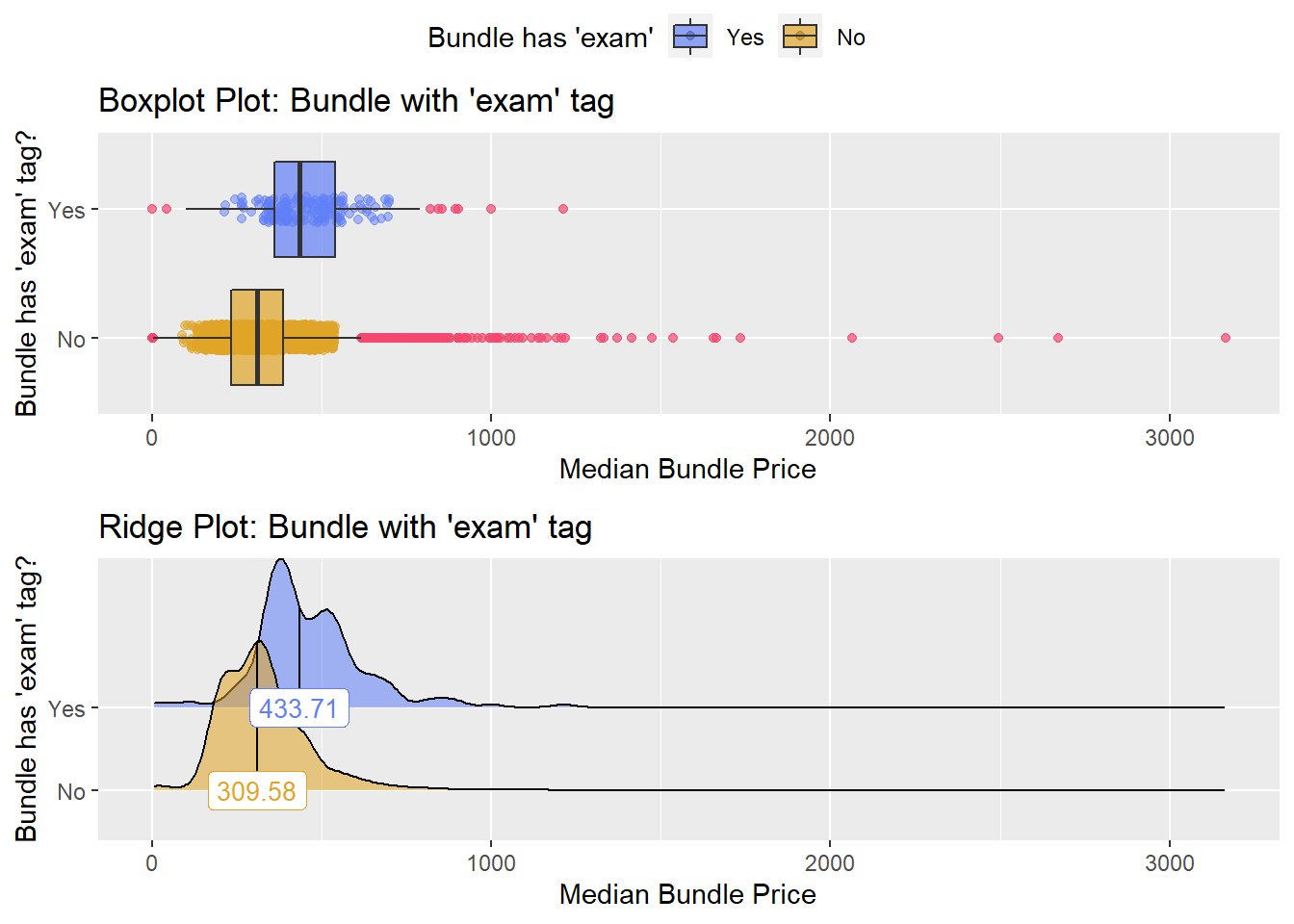
##
## $stat_tables
toe_nail2_clean <- toe_nail2_clean[radi_bun_t_exam==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.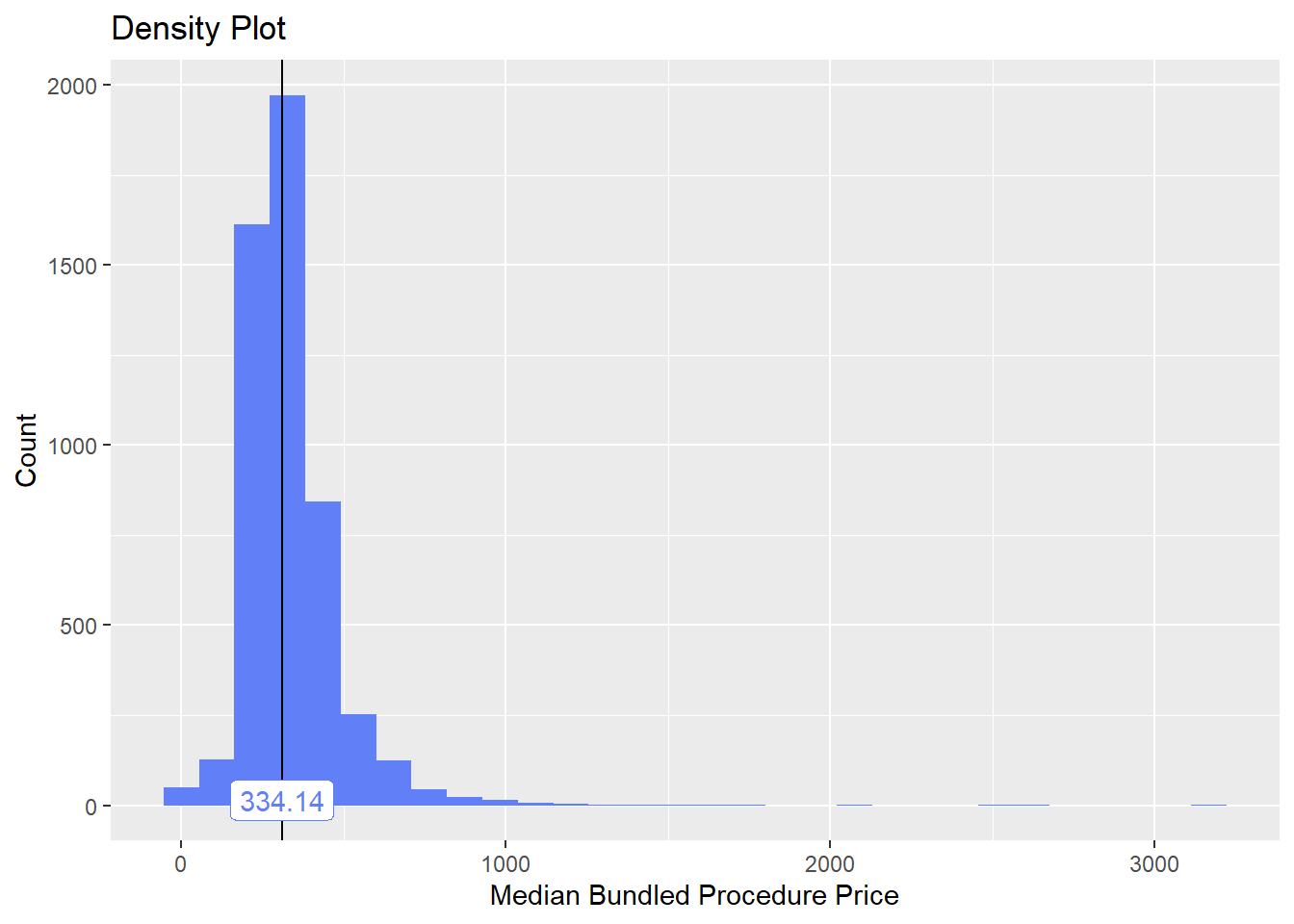
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 190 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.358
## 2 faci_bun_t_pat 0.351
## 3 faci_bun_t_office 0.351
## 4 path_bun_t_detect 0.128
## 5 path_bun_t_agent 0.126
## 6 path_bun_t_dna 0.126
## 7 faci_bun_t_est 0.124
## 8 medi_bun_t_hpv 0.110
## 9 medi_bun_t_evaluat 0.107
## 10 medi_bun_t_exercise 0.101
## # ... with 180 more rowstoe_nail2_clean %>% get_tag_density_information("path_bun_t_detect") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_detect"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 96## $dist_plots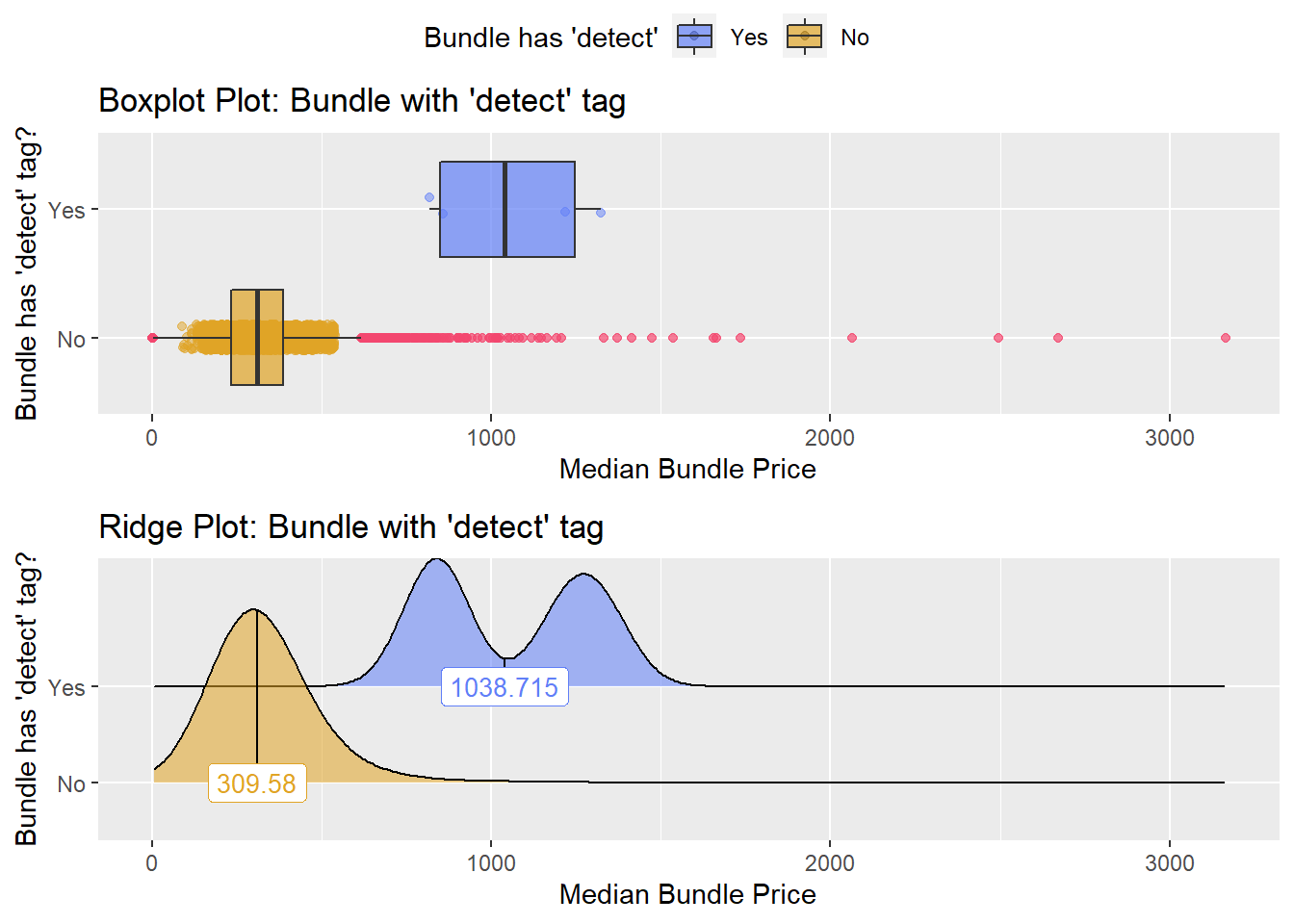
##
## $stat_tables
toe_nail2_clean <- toe_nail2_clean[path_bun_t_detect==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.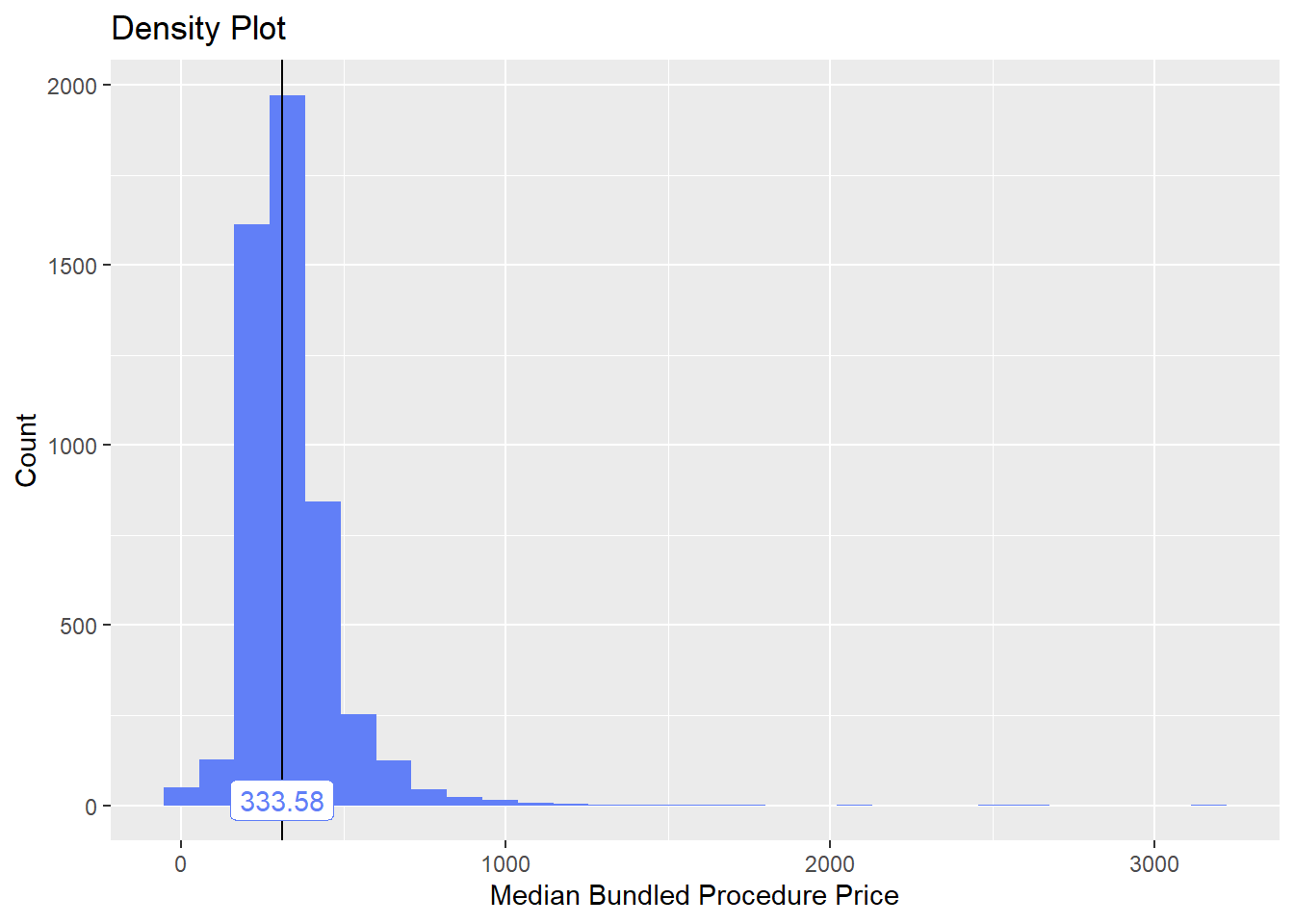
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 189 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.360
## 2 faci_bun_t_pat 0.353
## 3 faci_bun_t_office 0.352
## 4 faci_bun_t_est 0.127
## 5 medi_bun_t_hpv 0.111
## 6 medi_bun_t_evaluat 0.108
## 7 medi_bun_t_exercise 0.102
## 8 medi_bun_t_psytx 0.100
## 9 surg_bun_t_art 0.0978
## 10 medi_bun_t_therapeutic 0.0971
## # ... with 179 more rowstoe_nail2_clean %>% get_tag_density_information("medi_bun_t_hpv") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_hpv"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 73.6## $dist_plots
##
## $stat_tables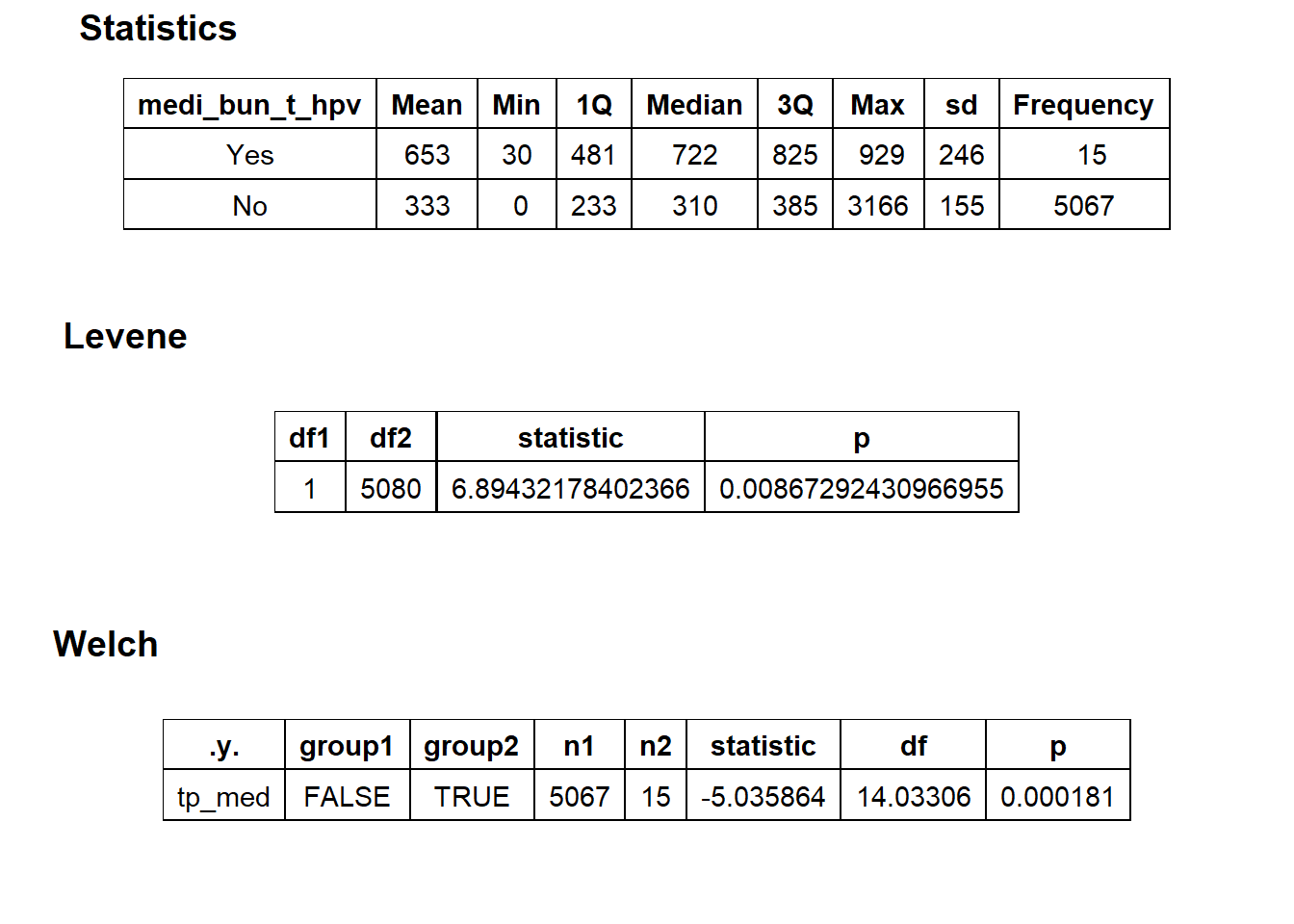
toe_nail2_clean <- toe_nail2_clean[medi_bun_t_hpv==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 188 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.363
## 2 faci_bun_t_pat 0.357
## 3 faci_bun_t_office 0.357
## 4 faci_bun_t_est 0.125
## 5 medi_bun_t_evaluat 0.109
## 6 medi_bun_t_exercise 0.103
## 7 medi_bun_t_psytx 0.102
## 8 surg_bun_t_art 0.0989
## 9 medi_bun_t_therapeutic 0.0986
## 10 surg_bun_t_arthro 0.0905
## # ... with 178 more rowstoe_nail2_clean %>% get_tag_density_information("medi_bun_t_evaluat") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_evaluat"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 82.8## $dist_plots
##
## $stat_tables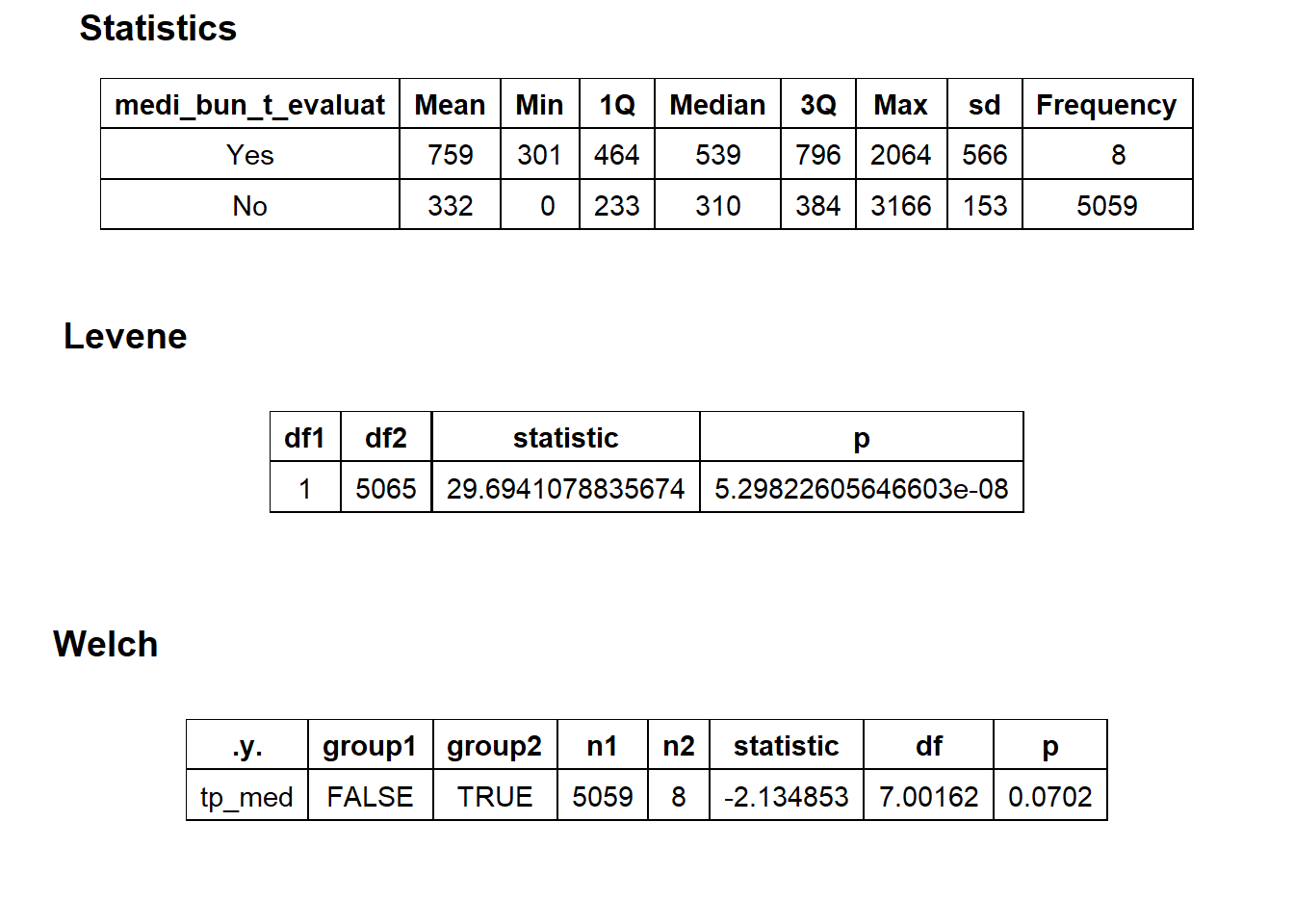
toe_nail2_clean <- toe_nail2_clean[medi_bun_t_evaluat==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 187 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.366
## 2 faci_bun_t_pat 0.360
## 3 faci_bun_t_office 0.360
## 4 faci_bun_t_est 0.128
## 5 surg_bun_t_art 0.100
## 6 surg_bun_t_arthro 0.0919
## 7 surg_bun_t_arthroscopy 0.0919
## 8 surg_bun_t_knee 0.0919
## 9 medi_bun_t_methylprednisolone 0.091
## 10 medi_bun_t_prednisolone 0.091
## # ... with 177 more rowstoe_nail2_clean %>% get_tag_density_information("surg_bun_t_art") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_art"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 130## $dist_plots
##
## $stat_tables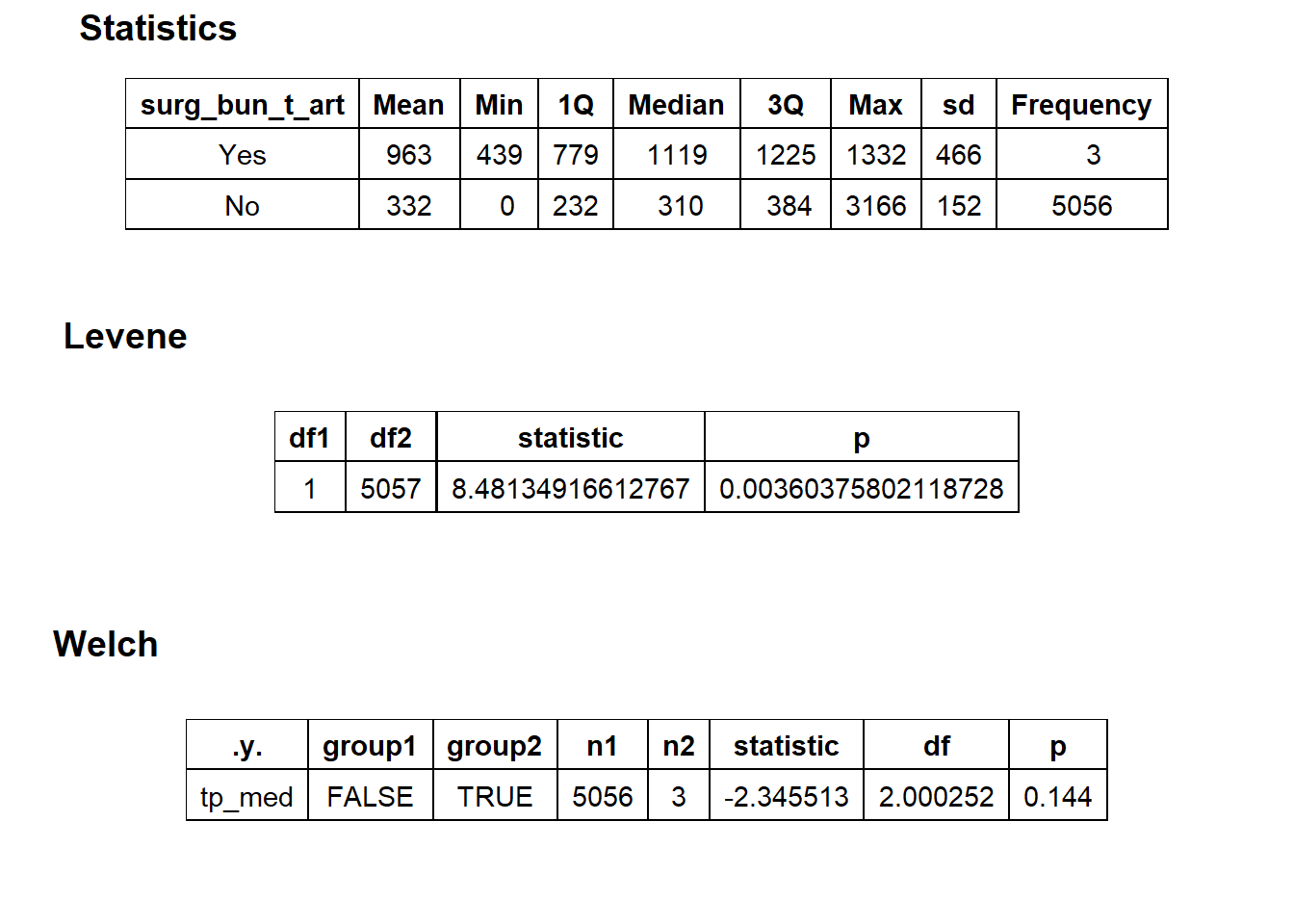
toe_nail2_clean <- toe_nail2_clean[surg_bun_t_art==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 180 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.369
## 2 faci_bun_t_pat 0.363
## 3 faci_bun_t_office 0.363
## 4 faci_bun_t_est 0.131
## 5 medi_bun_t_methylprednisolone 0.0918
## 6 medi_bun_t_prednisolone 0.0918
## 7 radi_bun_t_breast 0.09
## 8 radi_bun_t_mammo 0.0879
## 9 medi_bun_t_inject 0.0862
## 10 medi_bun_t_services 0.0857
## # ... with 170 more rowstoe_nail2_clean %>% get_tag_density_information("medi_bun_t_methylprednisolone") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_methylprednisolone"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 65.7## $dist_plots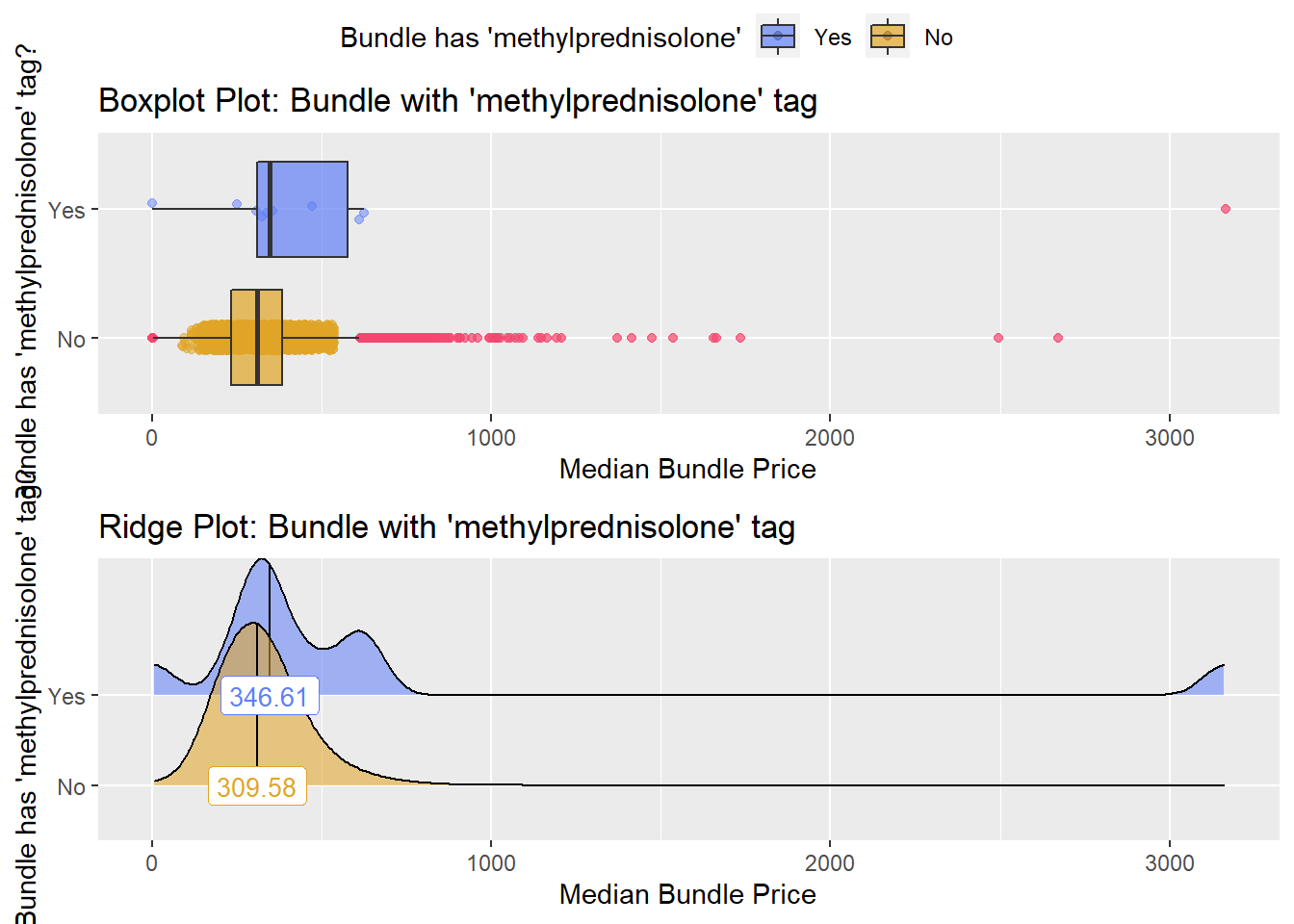
##
## $stat_tables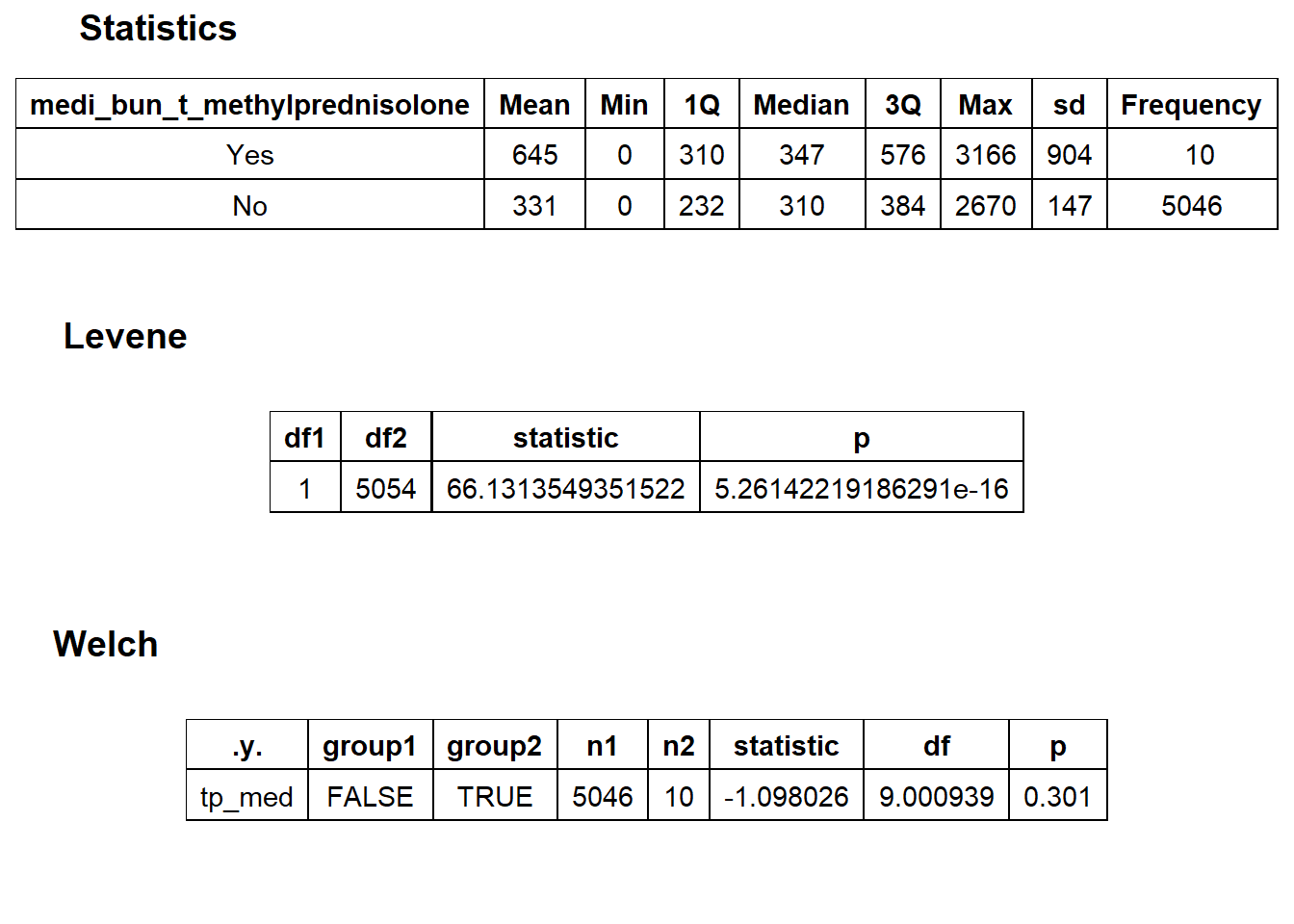
toe_nail2_clean <- toe_nail2_clean[medi_bun_t_methylprednisolone==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.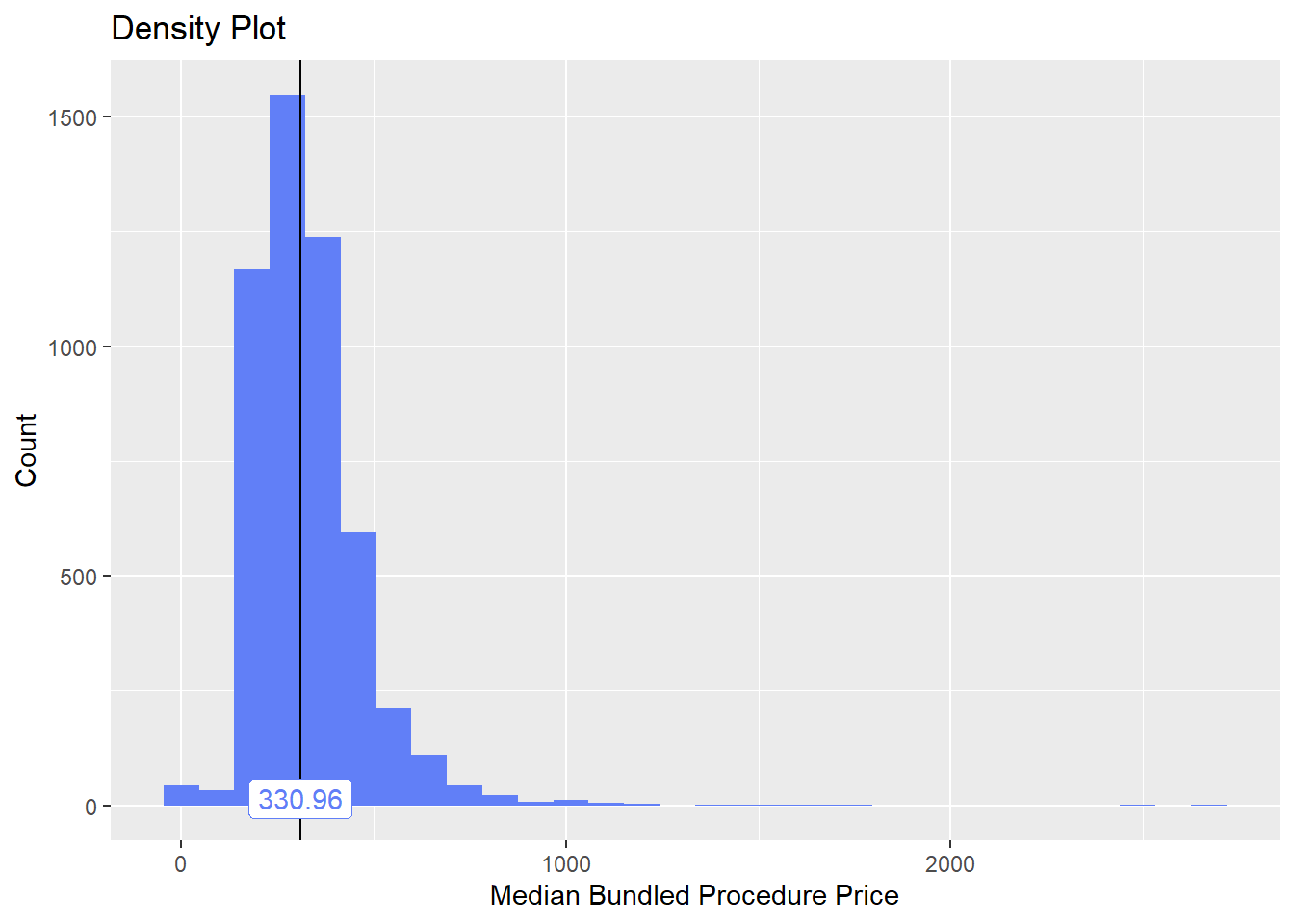
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 178 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.38
## 2 faci_bun_t_pat 0.373
## 3 faci_bun_t_office 0.373
## 4 faci_bun_t_est 0.129
## 5 radi_bun_t_breast 0.0935
## 6 radi_bun_t_mammo 0.0914
## 7 medi_bun_t_services 0.0891
## 8 medi_bun_t_psytx 0.0819
## 9 medi_bun_t_exercise 0.0795
## 10 medi_bun_t_eye_exam 0.0794
## # ... with 168 more rowstoe_nail2_clean %>% get_tag_density_information("radi_bun_t_breast") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ radi_bun_t_breast"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 38.7## $dist_plots
##
## $stat_tables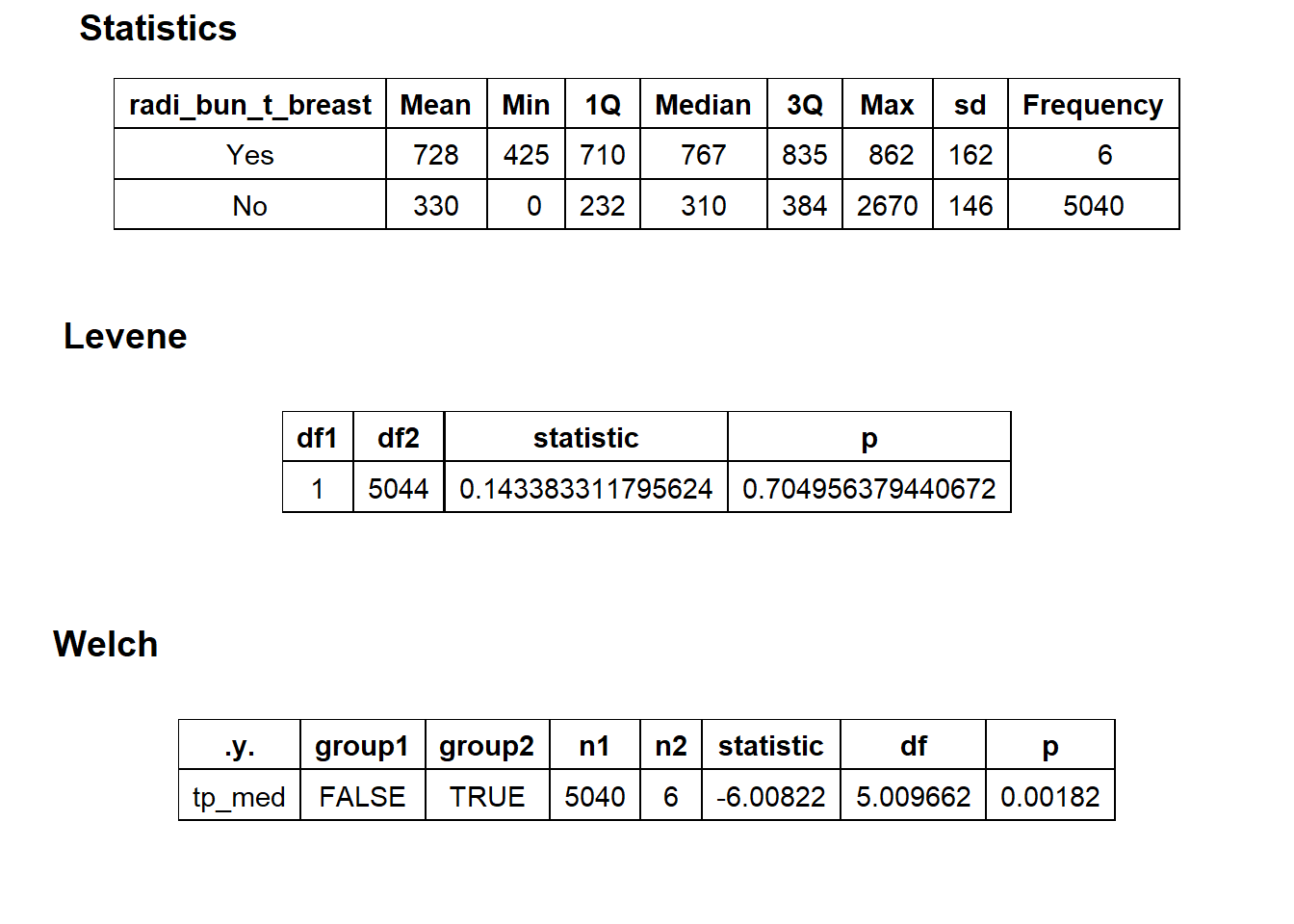
toe_nail2_clean <- toe_nail2_clean[radi_bun_t_breast==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 177 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.385
## 2 faci_bun_t_pat 0.378
## 3 faci_bun_t_office 0.378
## 4 faci_bun_t_est 0.130
## 5 medi_bun_t_services 0.0896
## 6 medi_bun_t_psytx 0.0827
## 7 medi_bun_t_exercise 0.0802
## 8 medi_bun_t_eye_exam 0.08
## 9 surg_bun_t_finger 0.0781
## 10 path_bun_t_assay 0.0762
## # ... with 167 more rowstoe_nail2_clean %>% get_tag_density_information("medi_bun_t_services") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_services"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 155## $dist_plots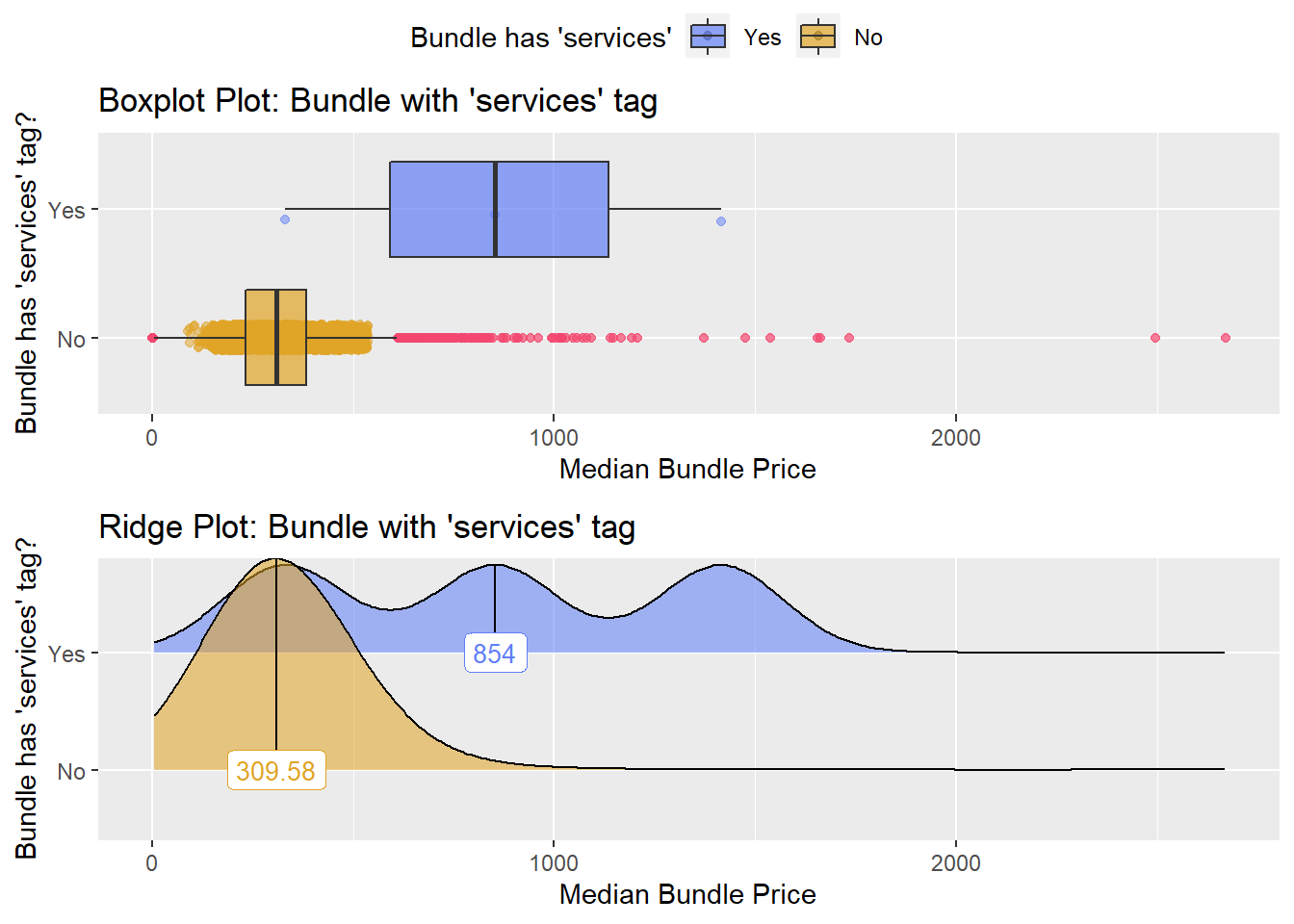
##
## $stat_tables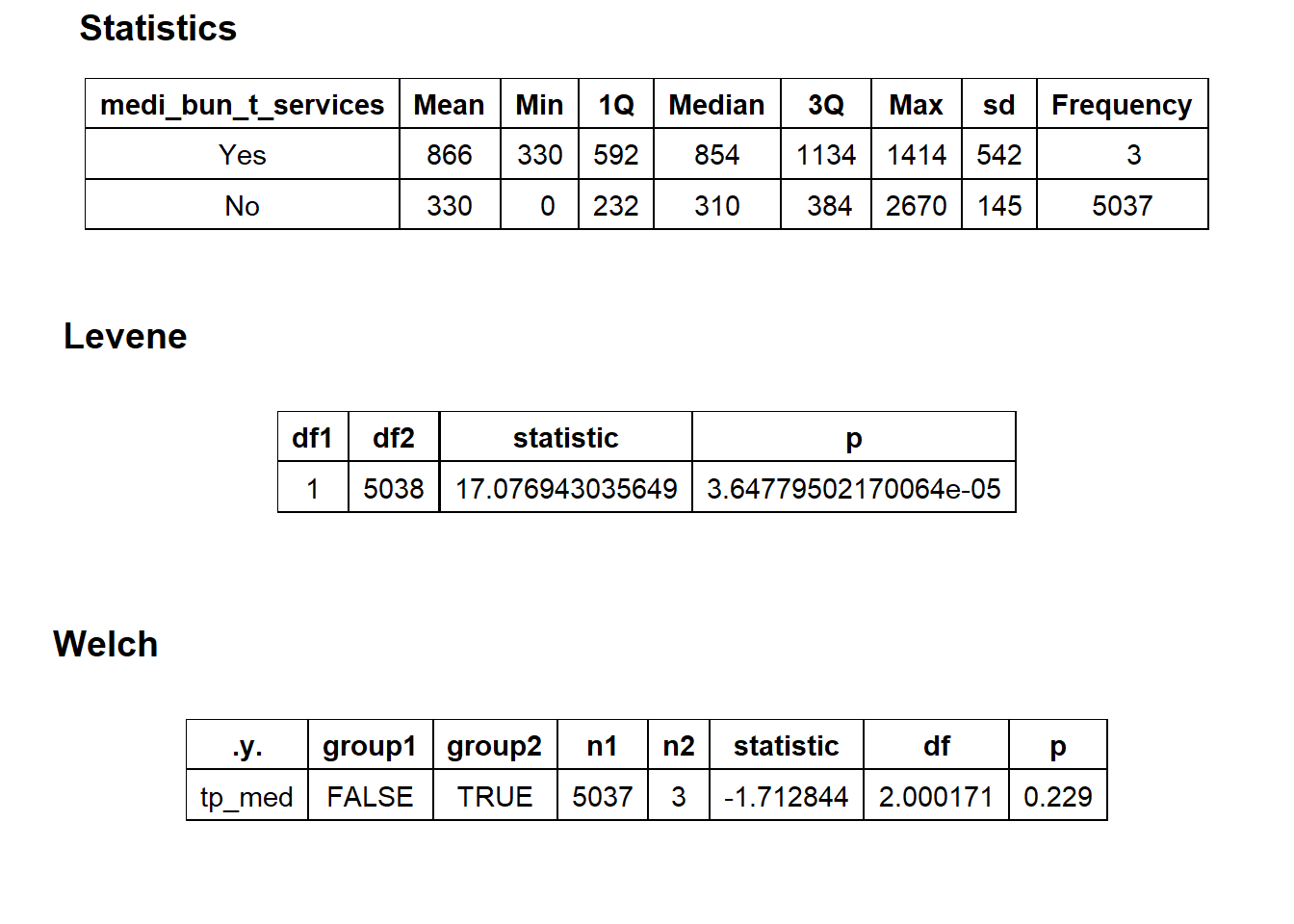
toe_nail2_clean <- toe_nail2_clean[medi_bun_t_services==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.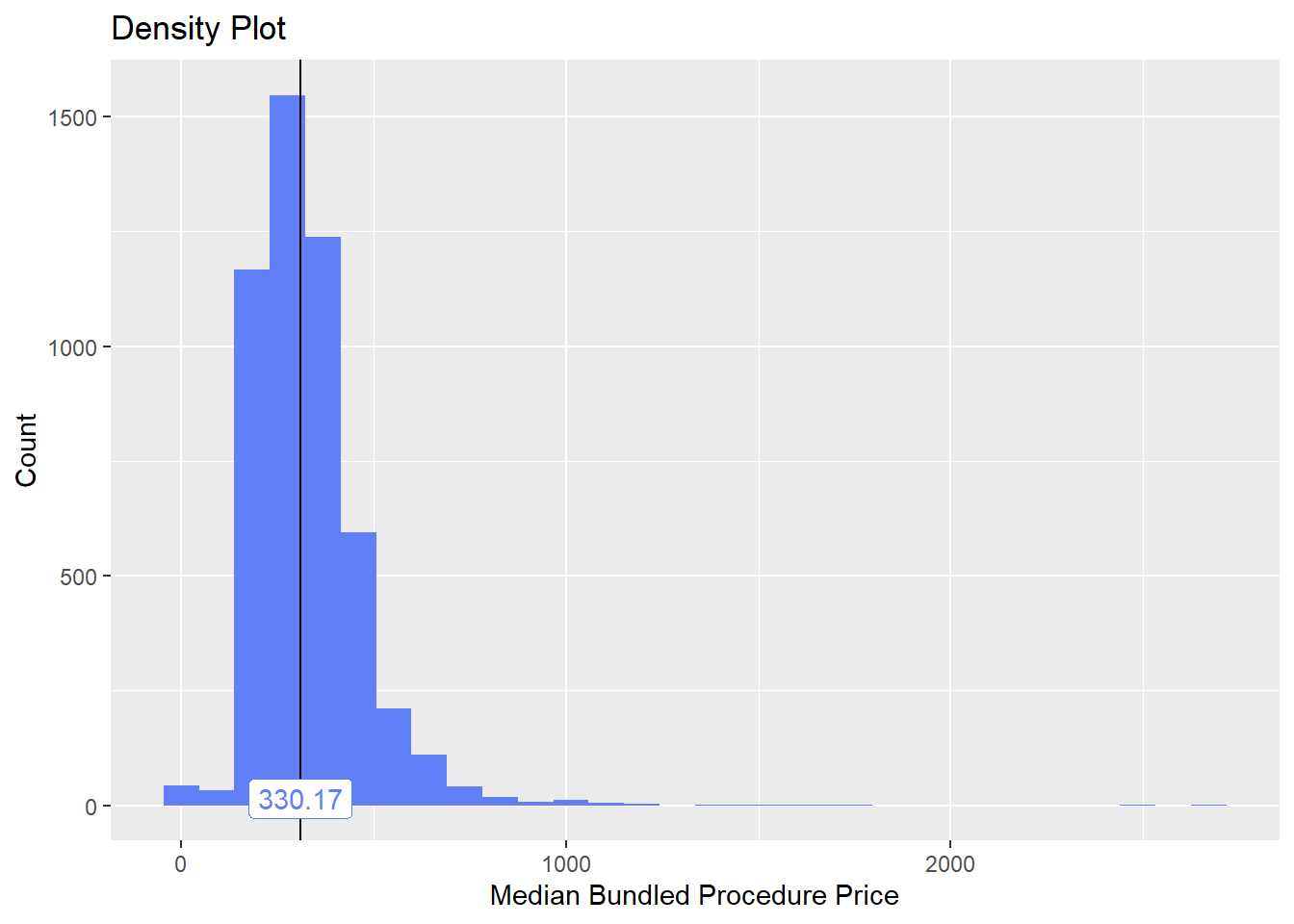
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 176 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.386
## 2 faci_bun_t_pat 0.379
## 3 faci_bun_t_office 0.379
## 4 faci_bun_t_est 0.132
## 5 medi_bun_t_psytx 0.0835
## 6 medi_bun_t_exercise 0.081
## 7 medi_bun_t_eye_exam 0.0807
## 8 surg_bun_t_finger 0.0786
## 9 path_bun_t_assay 0.0769
## 10 medi_bun_t_therapeutic 0.0766
## # ... with 166 more rowstoe_nail2_clean %>% get_tag_density_information("medi_bun_t_psytx") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_psytx"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 29.5## $dist_plots
##
## $stat_tables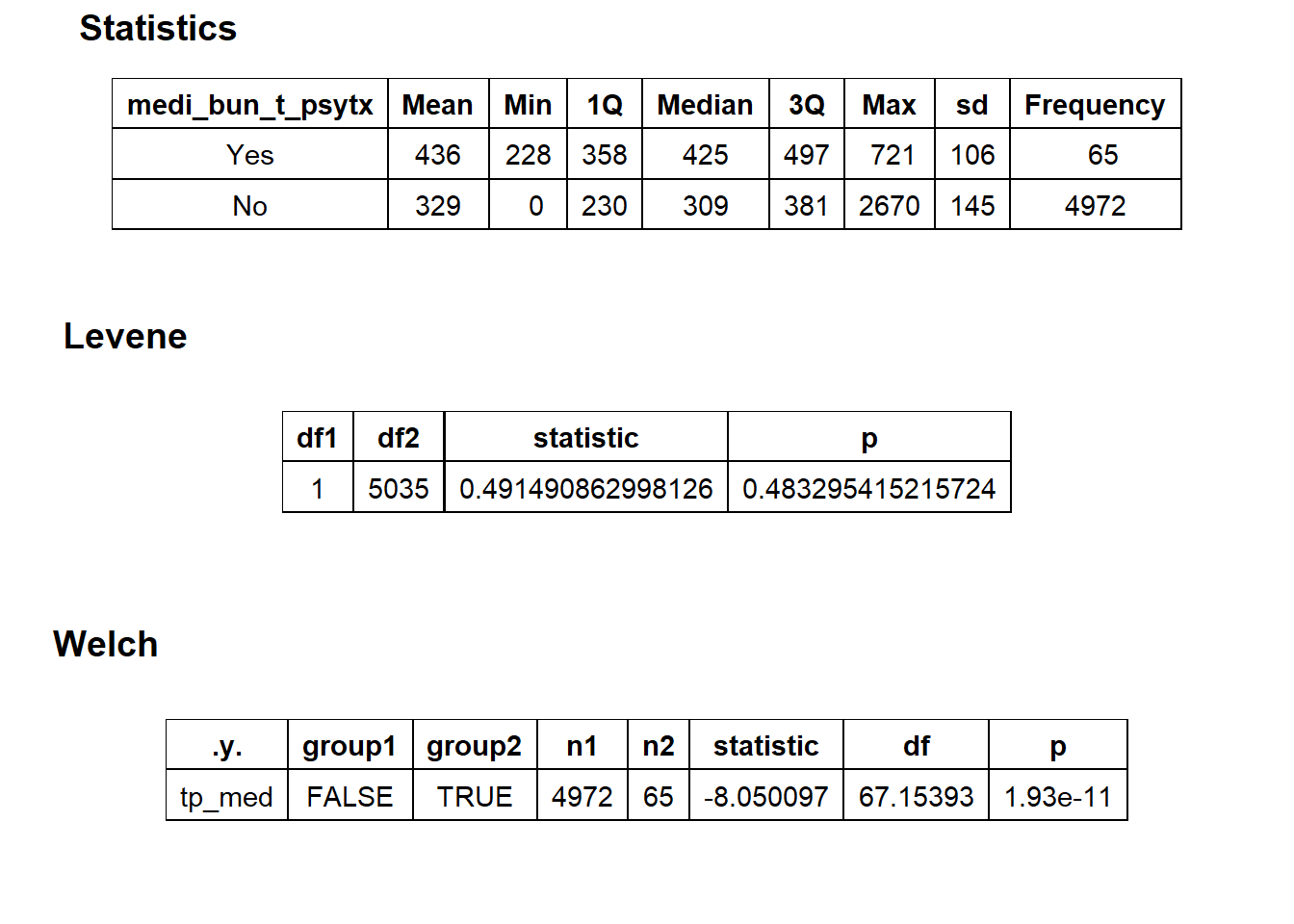
toe_nail2_clean <- toe_nail2_clean[medi_bun_t_psytx==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 174 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.385
## 2 faci_bun_t_pat 0.378
## 3 faci_bun_t_office 0.377
## 4 faci_bun_t_est 0.131
## 5 medi_bun_t_exercise 0.0824
## 6 medi_bun_t_eye_exam 0.082
## 7 surg_bun_t_finger 0.0793
## 8 path_bun_t_assay 0.0782
## 9 medi_bun_t_therapeutic 0.078
## 10 surg_bun_t_add-on 0.0762
## # ... with 164 more rowstoe_nail2_clean %>% get_tag_density_information("medi_bun_t_eye_exam") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_eye_exam"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 33.5## $dist_plots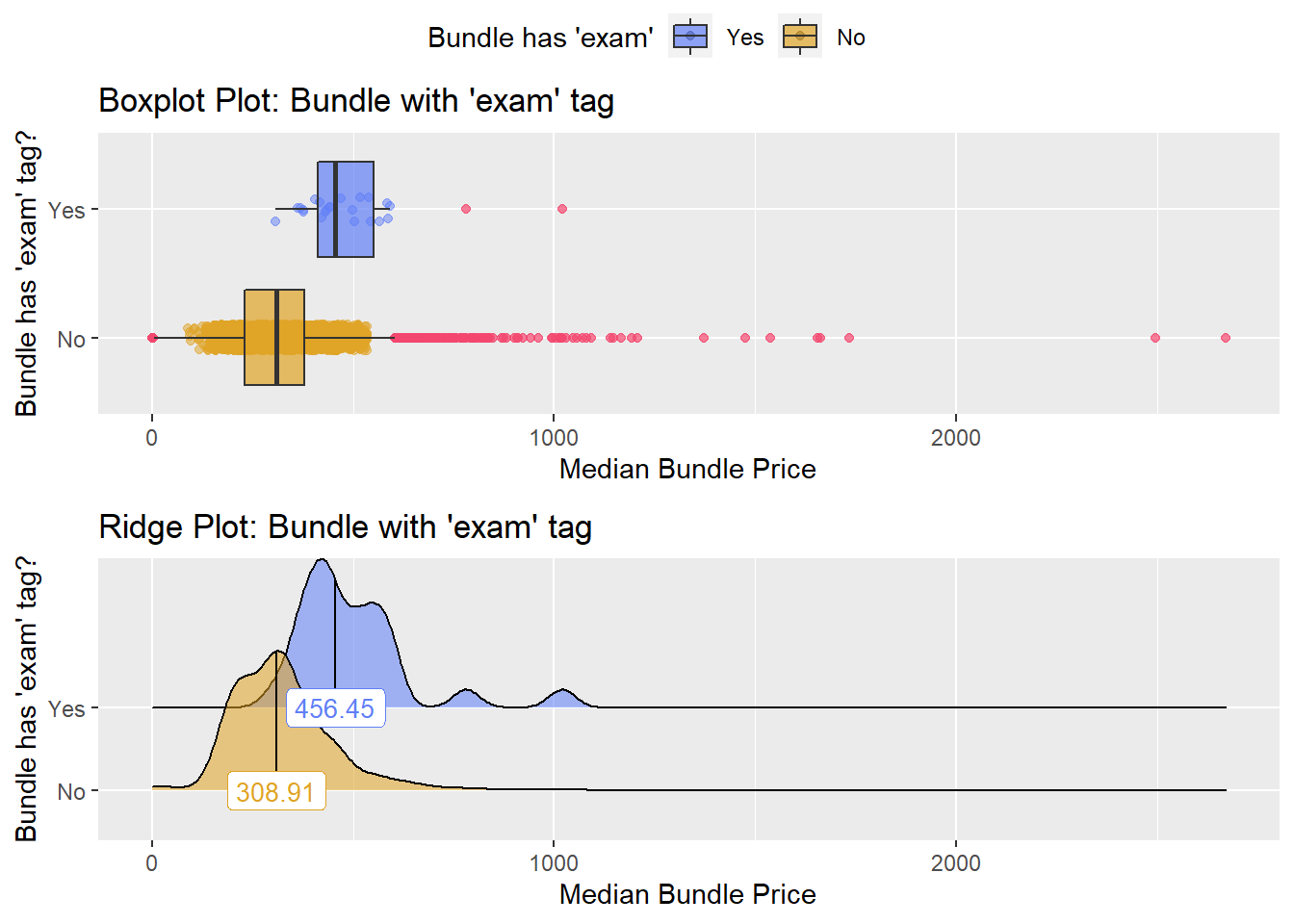
##
## $stat_tables
toe_nail2_clean <- toe_nail2_clean[medi_bun_t_eye_exam==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.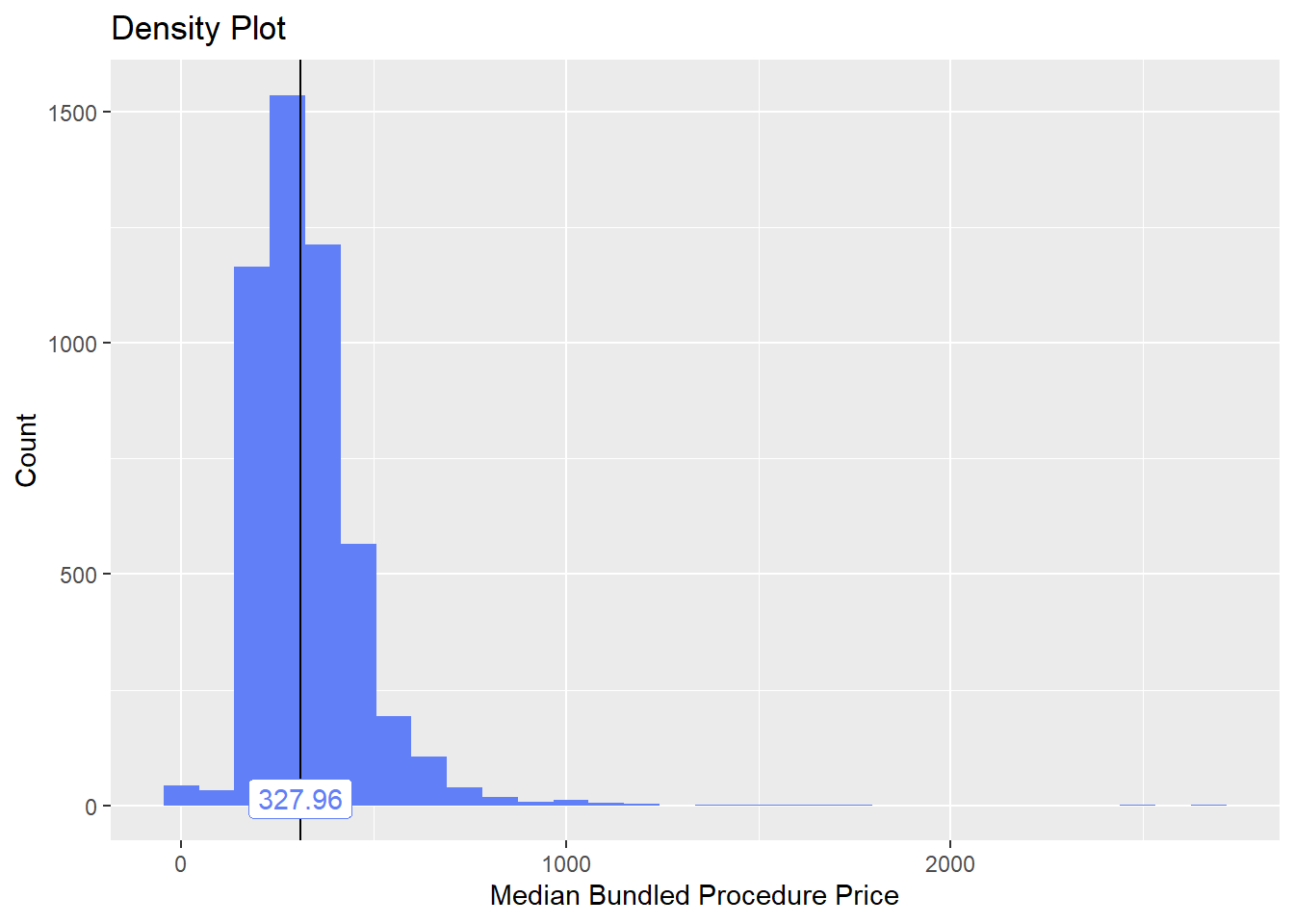
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 172 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.386
## 2 faci_bun_t_pat 0.378
## 3 faci_bun_t_office 0.378
## 4 faci_bun_t_est 0.129
## 5 medi_bun_t_exercise 0.0834
## 6 surg_bun_t_finger 0.0799
## 7 path_bun_t_assay 0.0791
## 8 medi_bun_t_therapeutic 0.079
## 9 surg_bun_t_add-on 0.0768
## 10 path_bun_t_test 0.0752
## # ... with 162 more rowstoe_nail2_clean %>% get_tag_density_information("medi_bun_t_exercise") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_exercise"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 38.9## $dist_plots
##
## $stat_tables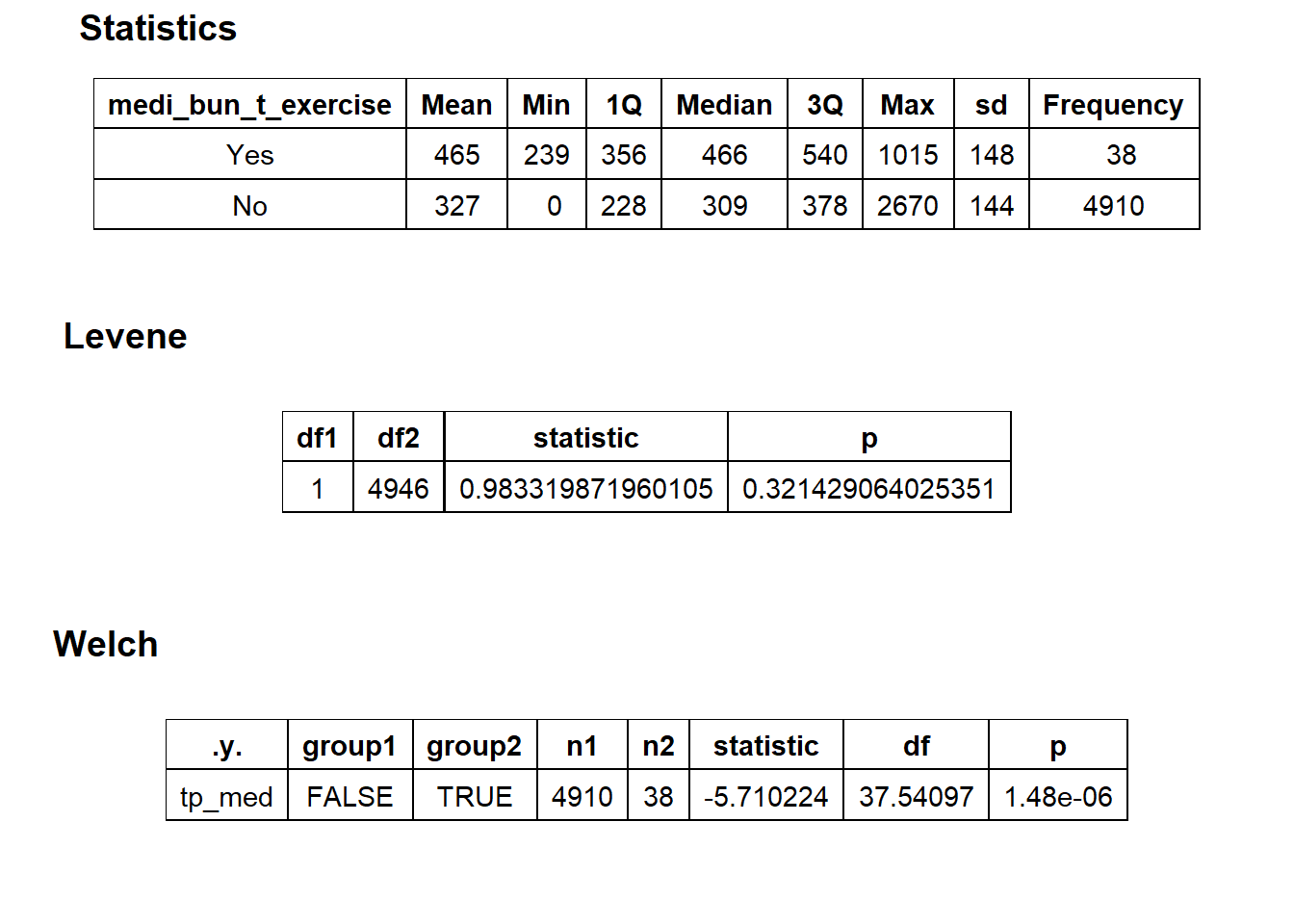
toe_nail2_clean <- toe_nail2_clean[medi_bun_t_exercise==F]toe_nail2_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.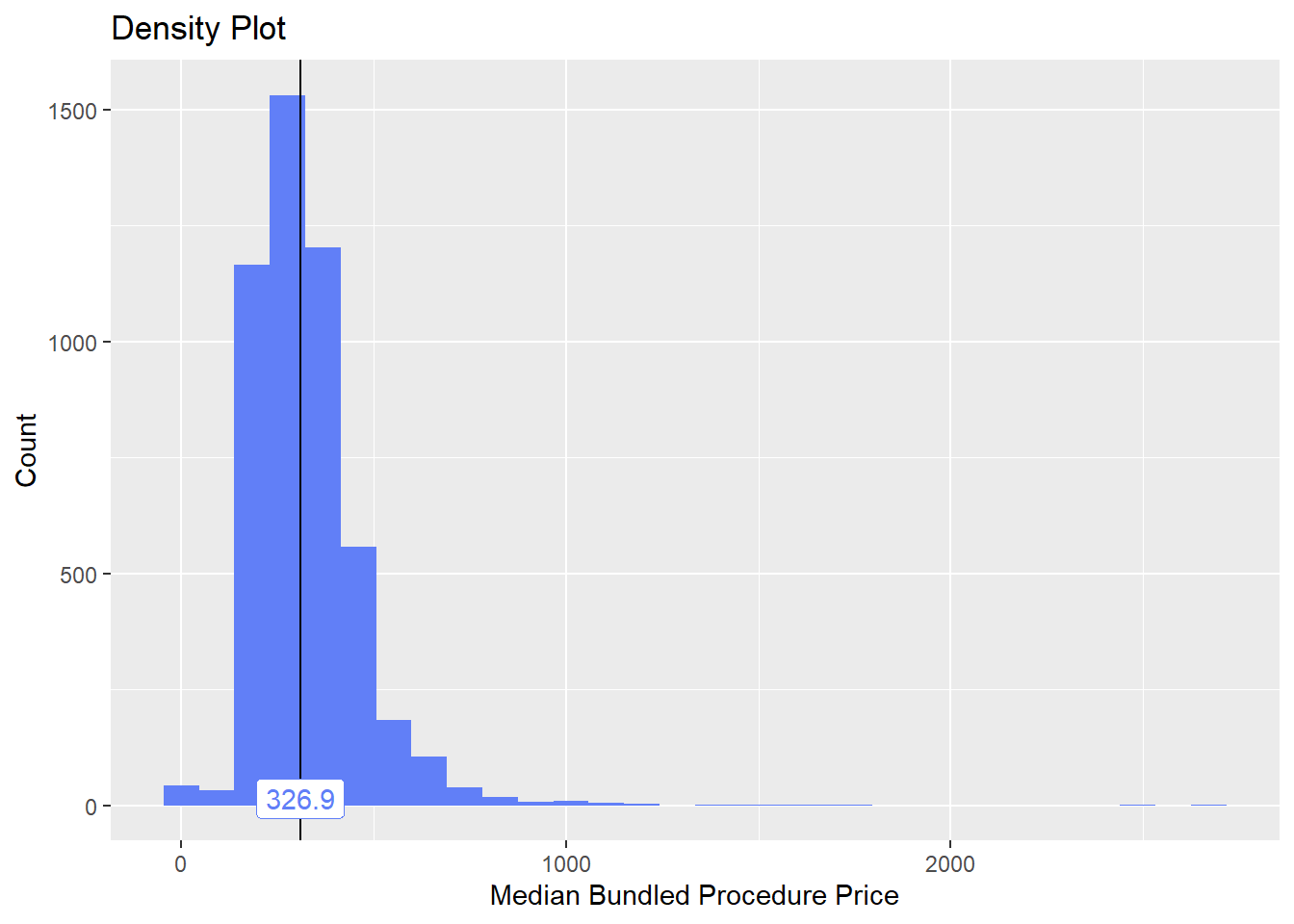
toe_nail2_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 171 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_visit 0.386
## 2 faci_bun_t_pat 0.378
## 3 faci_bun_t_office 0.378
## 4 faci_bun_t_est 0.130
## 5 surg_bun_t_finger 0.0806
## 6 path_bun_t_assay 0.0803
## 7 surg_bun_t_add-on 0.0776
## 8 medi_bun_t_boot 0.0766
## 9 path_bun_t_test 0.0726
## 10 radi_bun_t_without_dye 0.0724
## # ... with 161 more rowstoe_nail2_clean %>% get_tag_density_information("faci_bun_t_visit") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ faci_bun_t_visit"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 15.9## $dist_plots
##
## $stat_tables
toe_nail_w_visit <- toe_nail2_clean[faci_bun_t_visit==T]
toe_nail_n_visit <- toe_nail2_clean[faci_bun_t_visit==F]##toe_nail_w_visit
toe_nail_w_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.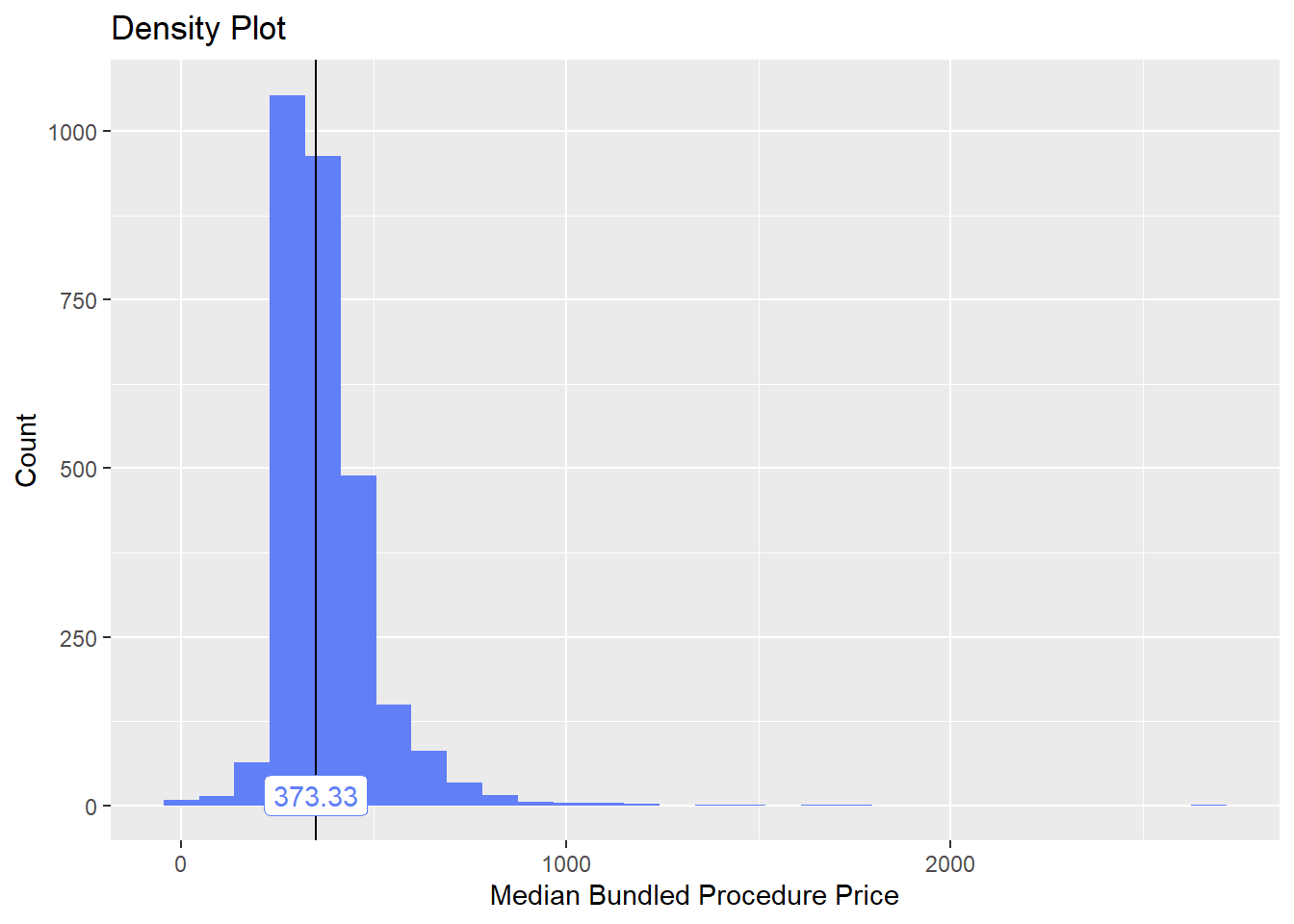
toe_nail_w_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 162 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_add-on 0.148
## 2 surg_bun_t_finger 0.107
## 3 surg_bun_t_duct 0.0894
## 4 surg_bun_t_sperm 0.0894
## 5 path_bun_t_vitamin 0.0891
## 6 path_bun_t_assay 0.089
## 7 path_bun_t_progesterone 0.0881
## 8 path_bun_t_sex 0.0881
## 9 radi_bun_t_without_dye 0.0875
## 10 faci_bun_t_est 0.0836
## # ... with 152 more rowstoe_nail_w_visit[`surg_bun_t_add-on` ==T] %>% nrow()## [1] 2toe_nail_w_visit <- toe_nail_w_visit[`surg_bun_t_add-on` ==F] toe_nail_w_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.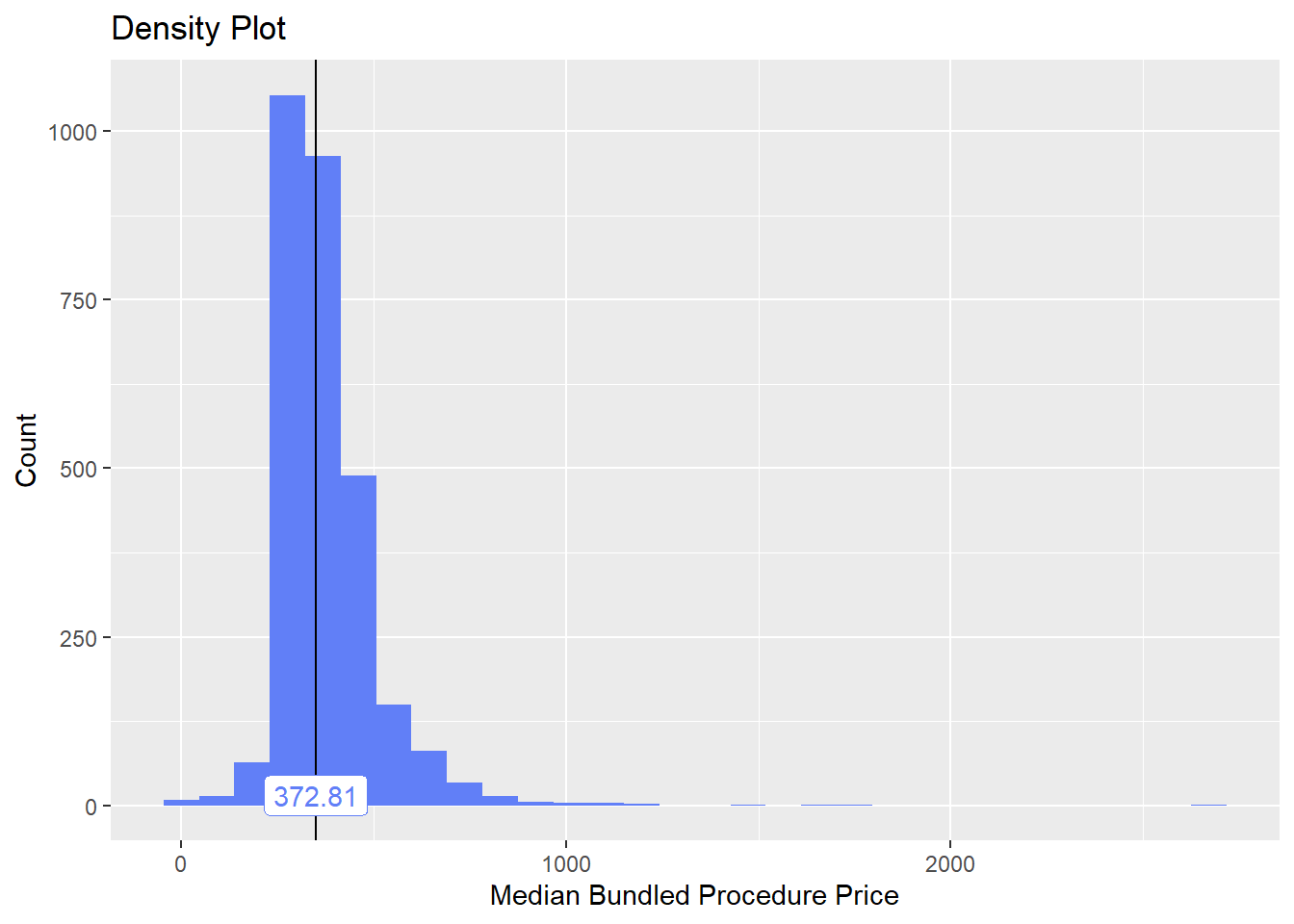
toe_nail_w_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 160 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_finger 0.108
## 2 surg_bun_t_duct 0.0905
## 3 surg_bun_t_sperm 0.0905
## 4 path_bun_t_assay 0.0905
## 5 path_bun_t_vitamin 0.0903
## 6 faci_bun_t_est 0.0894
## 7 path_bun_t_progesterone 0.0893
## 8 path_bun_t_sex 0.0893
## 9 radi_bun_t_without_dye 0.0888
## 10 duration_mean 0.0831
## # ... with 150 more rowstoe_nail_w_visit[surg_bun_t_finger==T] %>% nrow()## [1] 1toe_nail_w_visit <- toe_nail_w_visit[surg_bun_t_finger==F]toe_nail_w_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.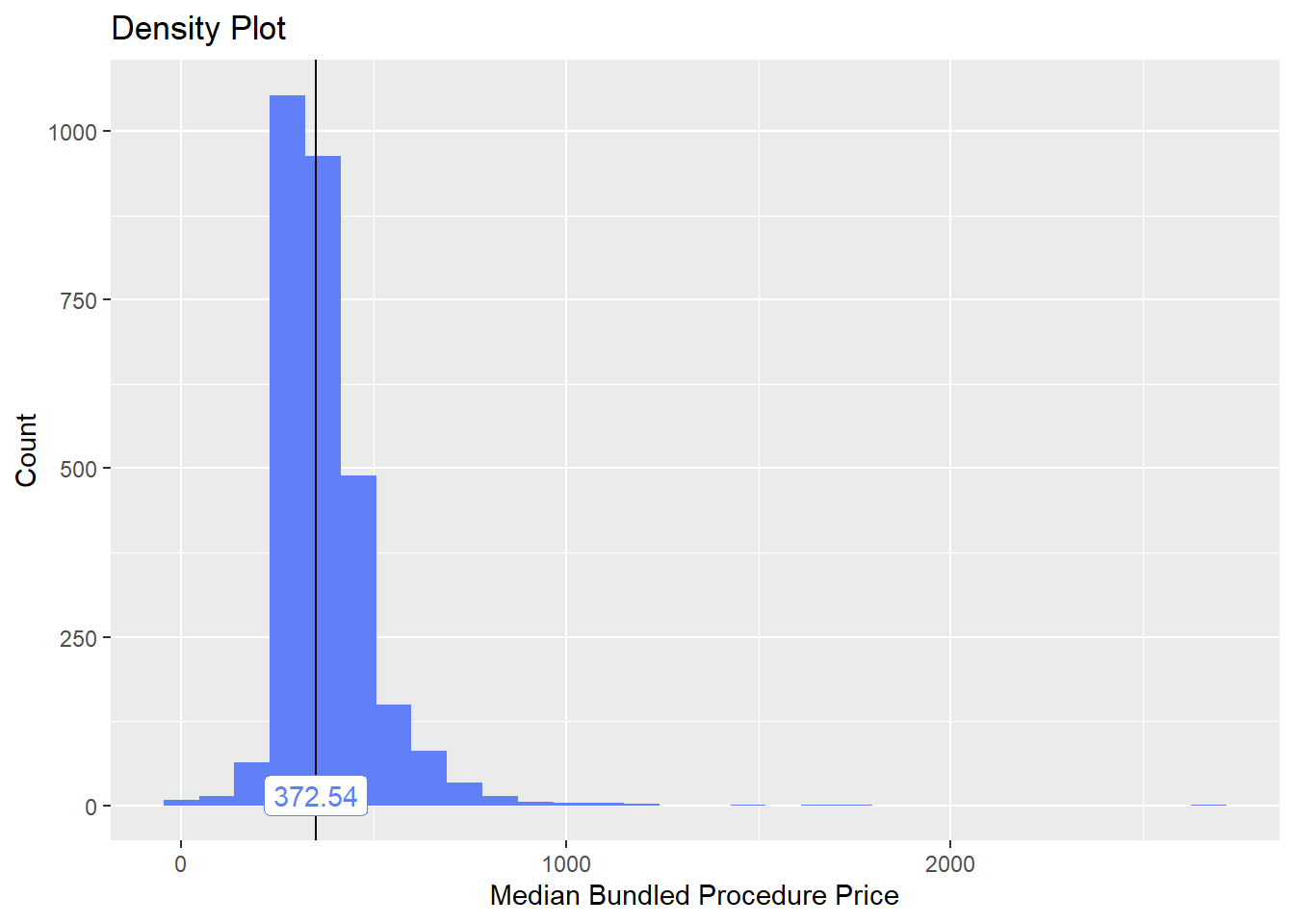
toe_nail_w_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 159 x 2
## name correlation
## <chr> <dbl>
## 1 path_bun_t_assay 0.0913
## 2 surg_bun_t_duct 0.0911
## 3 surg_bun_t_sperm 0.0911
## 4 path_bun_t_vitamin 0.0909
## 5 path_bun_t_progesterone 0.0899
## 6 path_bun_t_sex 0.0899
## 7 radi_bun_t_without_dye 0.0894
## 8 faci_bun_t_est 0.0882
## 9 duration_mean 0.0837
## 10 duration_max 0.0837
## # ... with 149 more rowstoe_nail_w_visit %>% get_tag_density_information("path_bun_t_assay") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_assay"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 58.7## $dist_plots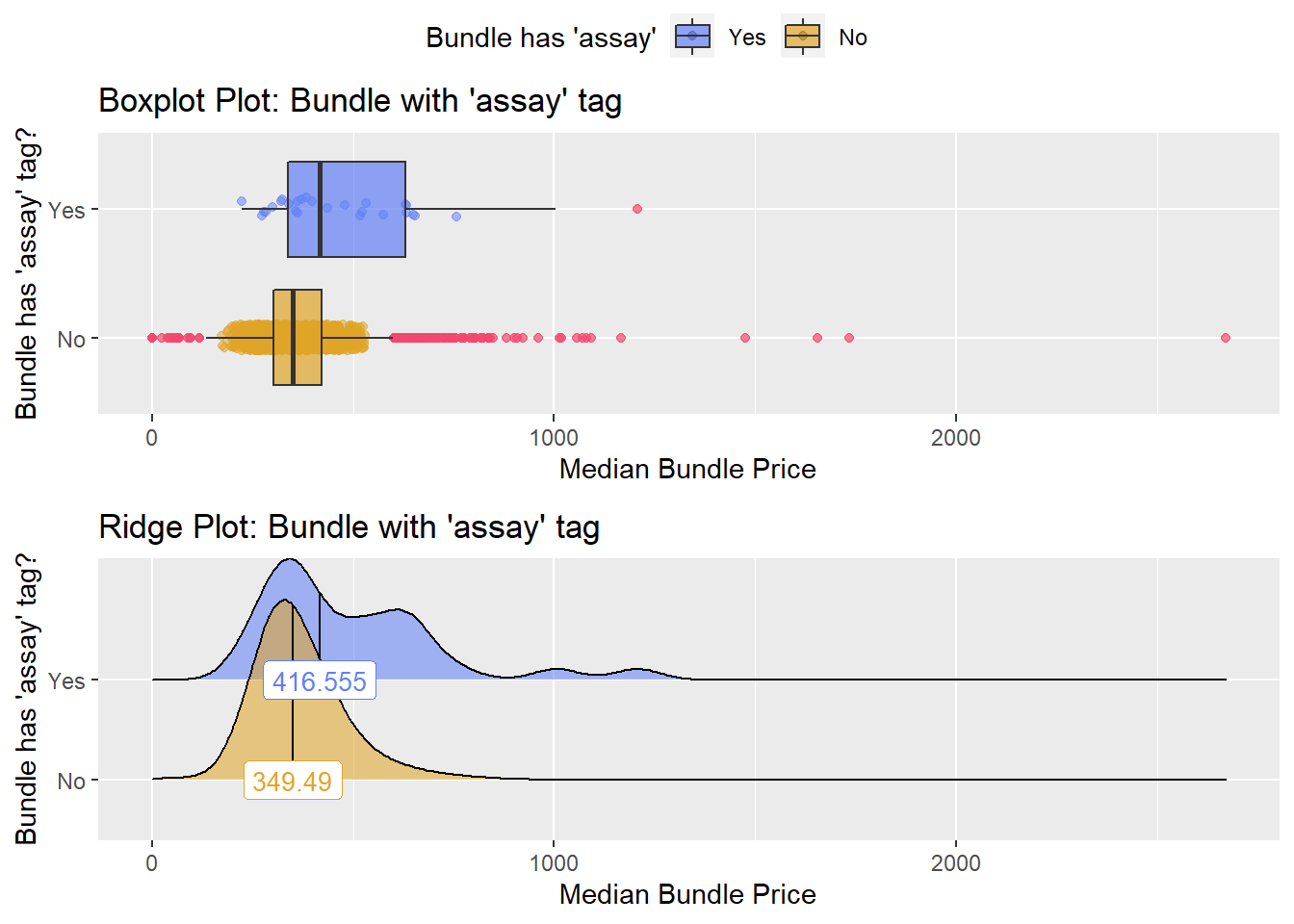
##
## $stat_tables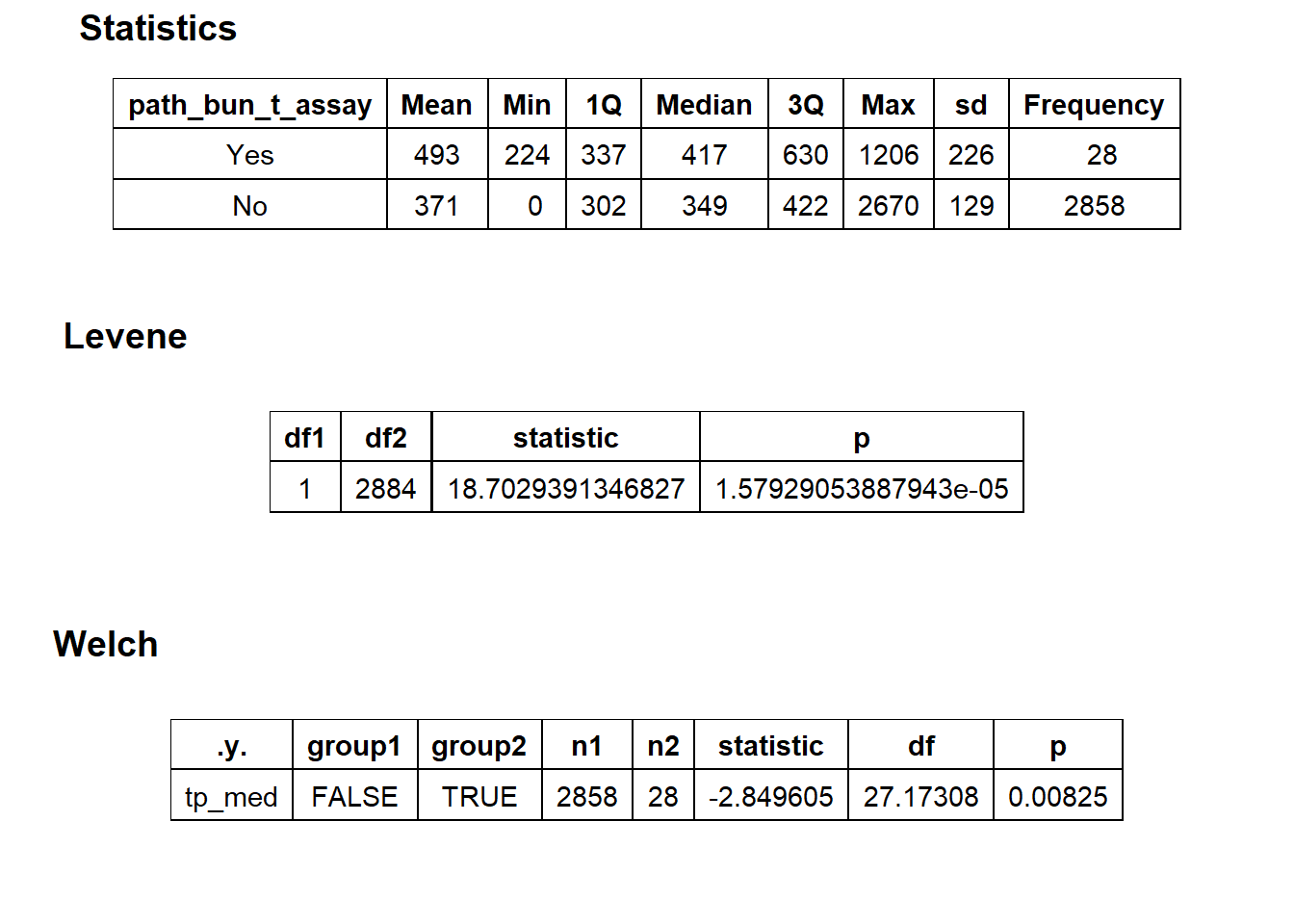
toe_nail_w_visit <- toe_nail_w_visit[path_bun_t_assay==F]toe_nail_w_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail_w_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 140 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_duct 0.093
## 2 surg_bun_t_sperm 0.093
## 3 faci_bun_t_est 0.093
## 4 radi_bun_t_without_dye 0.0914
## 5 duration_max 0.0854
## 6 duration_mean 0.0854
## 7 path_bun_t_cytopath 0.0744
## 8 path_bun_t_fluid 0.0724
## 9 radi_bun_t_mri 0.0678
## 10 surg_bun_t_interlaminar 0.0652
## # ... with 130 more rows# toe_nail_w_visit %>% get_tag_density_information("surg_bun_t_duct") %>% print()
# only one observationtoe_nail_w_visit <- toe_nail_w_visit[path_bun_t_assay==F]toe_nail_w_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail_w_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 140 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_duct 0.093
## 2 surg_bun_t_sperm 0.093
## 3 faci_bun_t_est 0.093
## 4 radi_bun_t_without_dye 0.0914
## 5 duration_max 0.0854
## 6 duration_mean 0.0854
## 7 path_bun_t_cytopath 0.0744
## 8 path_bun_t_fluid 0.0724
## 9 radi_bun_t_mri 0.0678
## 10 surg_bun_t_interlaminar 0.0652
## # ... with 130 more rows# toe_nail_w_visit %>% get_tag_density_information("surg_bun_t_sperm") %>% print()
# only one observationtoe_nail_w_visit <- toe_nail_w_visit[surg_bun_t_sperm==F]toe_nail_w_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail_w_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 138 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_est 0.0919
## 2 radi_bun_t_without_dye 0.0919
## 3 duration_mean 0.0858
## 4 duration_max 0.0858
## 5 path_bun_t_cytopath 0.0747
## 6 path_bun_t_fluid 0.0727
## 7 radi_bun_t_mri 0.0681
## 8 surg_bun_t_interlaminar 0.0655
## 9 surg_bun_t_lamina 0.0655
## 10 surg_bun_t_njx 0.0655
## # ... with 128 more rowstoe_nail_w_visit %>% get_tag_density_information("path_bun_t_cytopath") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_cytopath"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 46.1## $dist_plots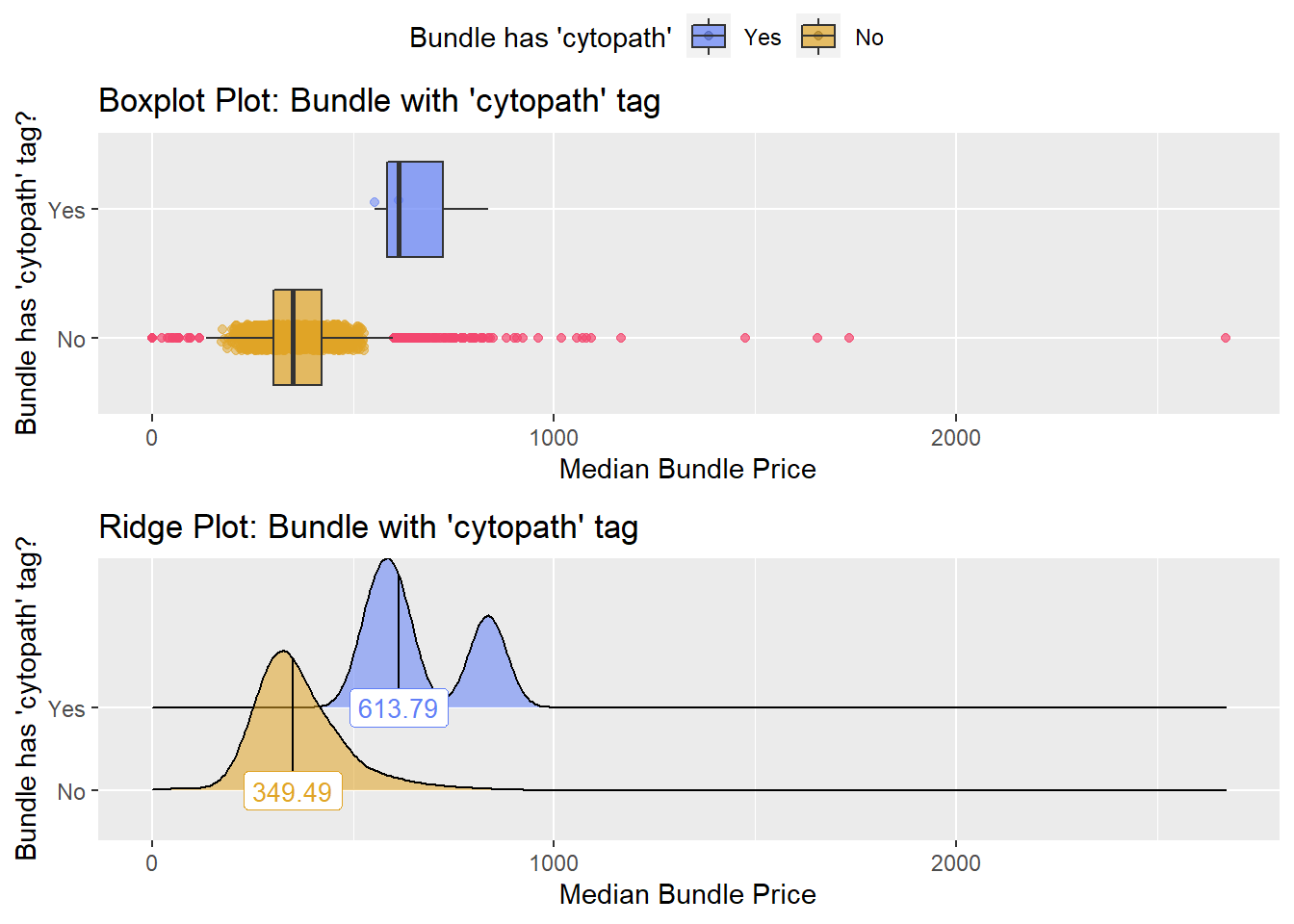
##
## $stat_tables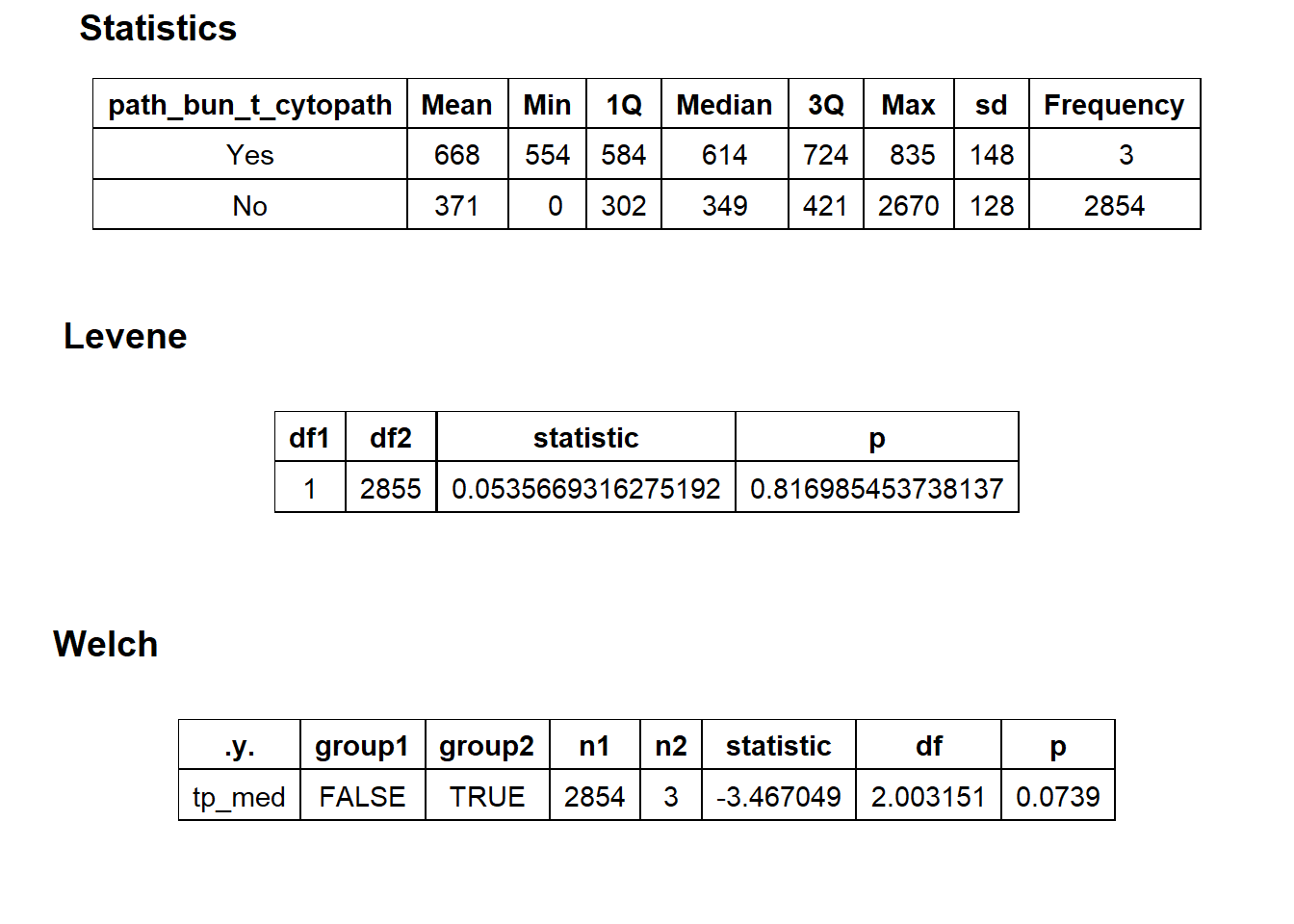
# only one observationtoe_nail_w_visit <- toe_nail_w_visit[path_bun_t_cytopath==F]toe_nail_w_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.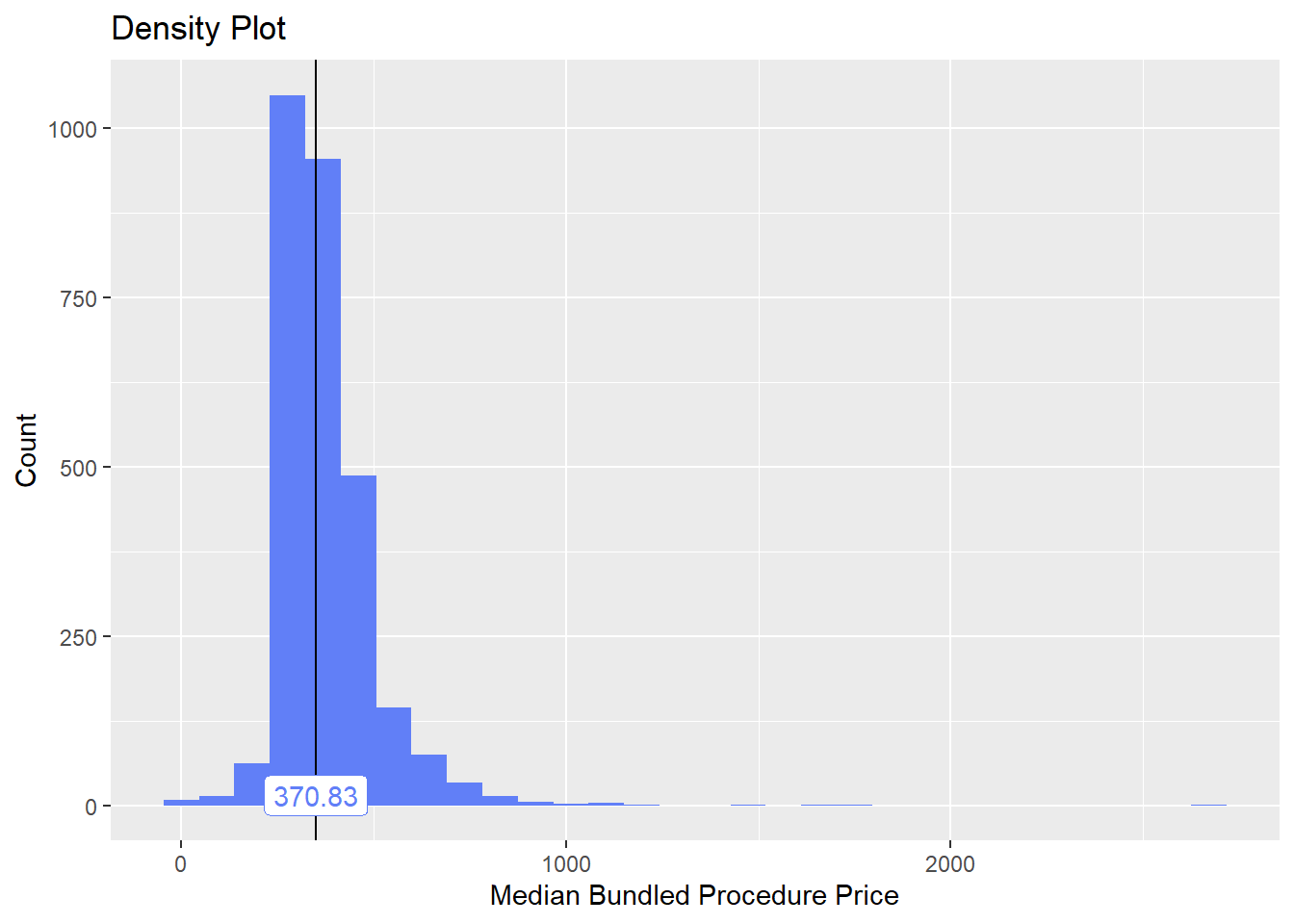
toe_nail_w_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 136 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_est 0.0952
## 2 radi_bun_t_without_dye 0.0923
## 3 duration_max 0.0861
## 4 duration_mean 0.0861
## 5 radi_bun_t_mri 0.0684
## 6 surg_bun_t_interlaminar 0.0658
## 7 surg_bun_t_lamina 0.0658
## 8 surg_bun_t_njx 0.0658
## 9 medi_bun_t_inject 0.0638
## 10 radi_bun_t_ct 0.0621
## # ... with 126 more rowstoe_nail_w_visit %>% get_tag_density_information("radi_bun_t_mri") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ radi_bun_t_mri"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 48.9## $dist_plots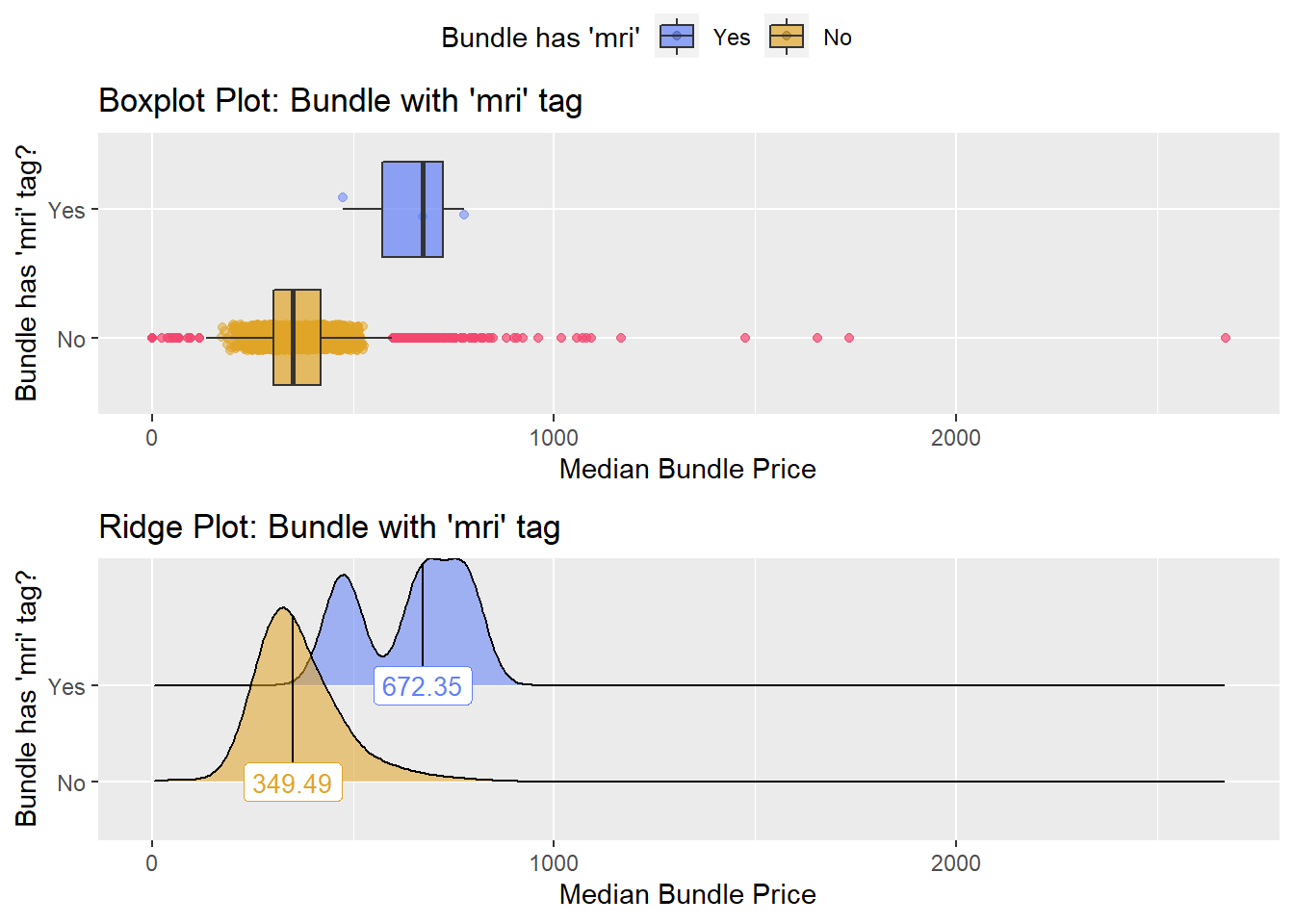
##
## $stat_tables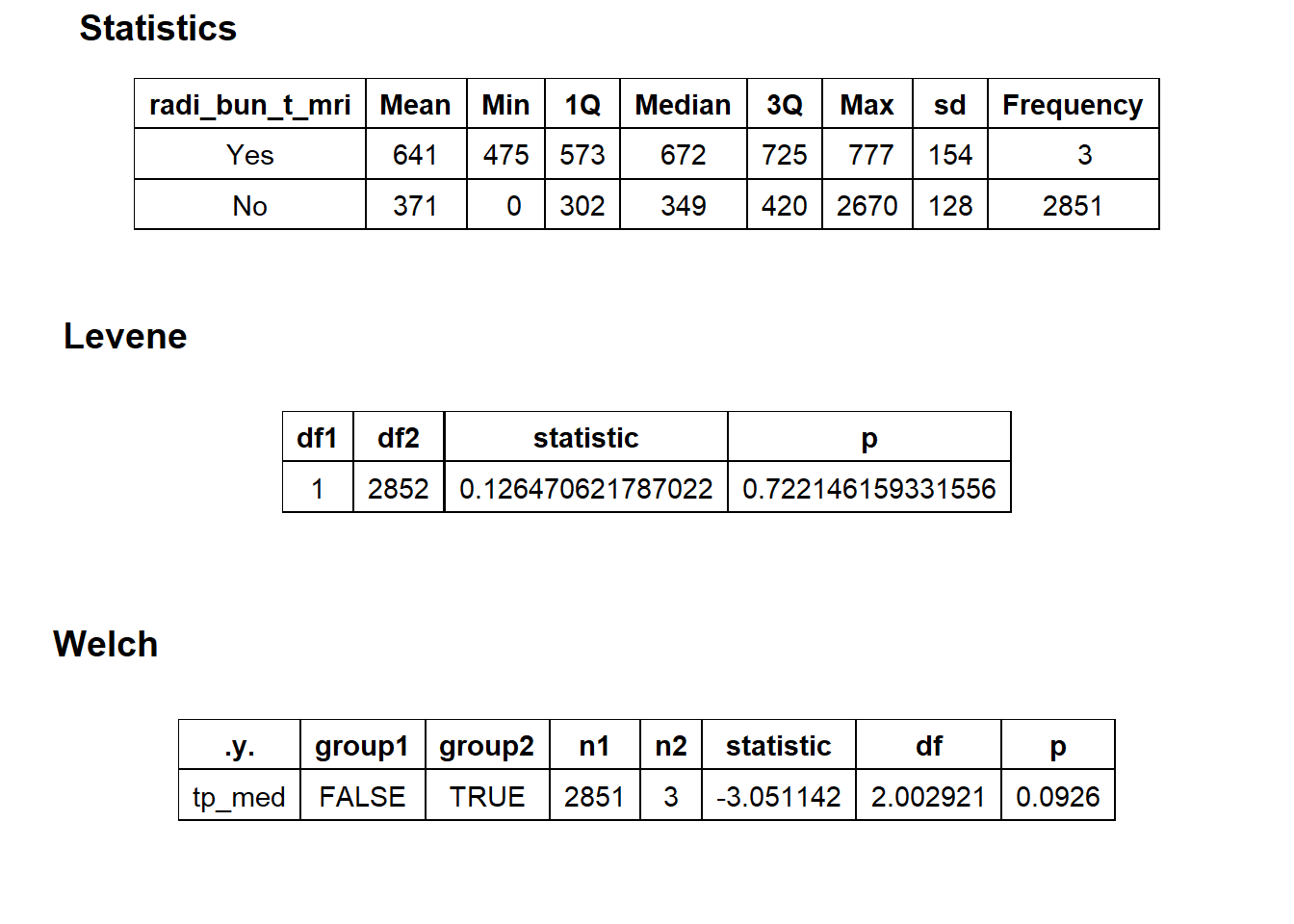
# only one observationtoe_nail_w_visit <- toe_nail_w_visit[radi_bun_t_mri==F]toe_nail_w_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail_w_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 132 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_est 0.0982
## 2 duration_mean 0.0864
## 3 duration_max 0.0864
## 4 surg_bun_t_interlaminar 0.066
## 5 surg_bun_t_lamina 0.066
## 6 surg_bun_t_njx 0.066
## 7 medi_bun_t_inject 0.0642
## 8 radi_bun_t_ct 0.0623
## 9 radi_bun_t_without_dye 0.0623
## 10 medi_bun_t_manual 0.0618
## # ... with 122 more rows# toe_nail_w_visit %>% get_tag_density_information("surg_bun_t_interlaminar") %>% print()
# only one observationtoe_nail_w_visit <- toe_nail_w_visit[surg_bun_t_interlaminar==F]toe_nail_w_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.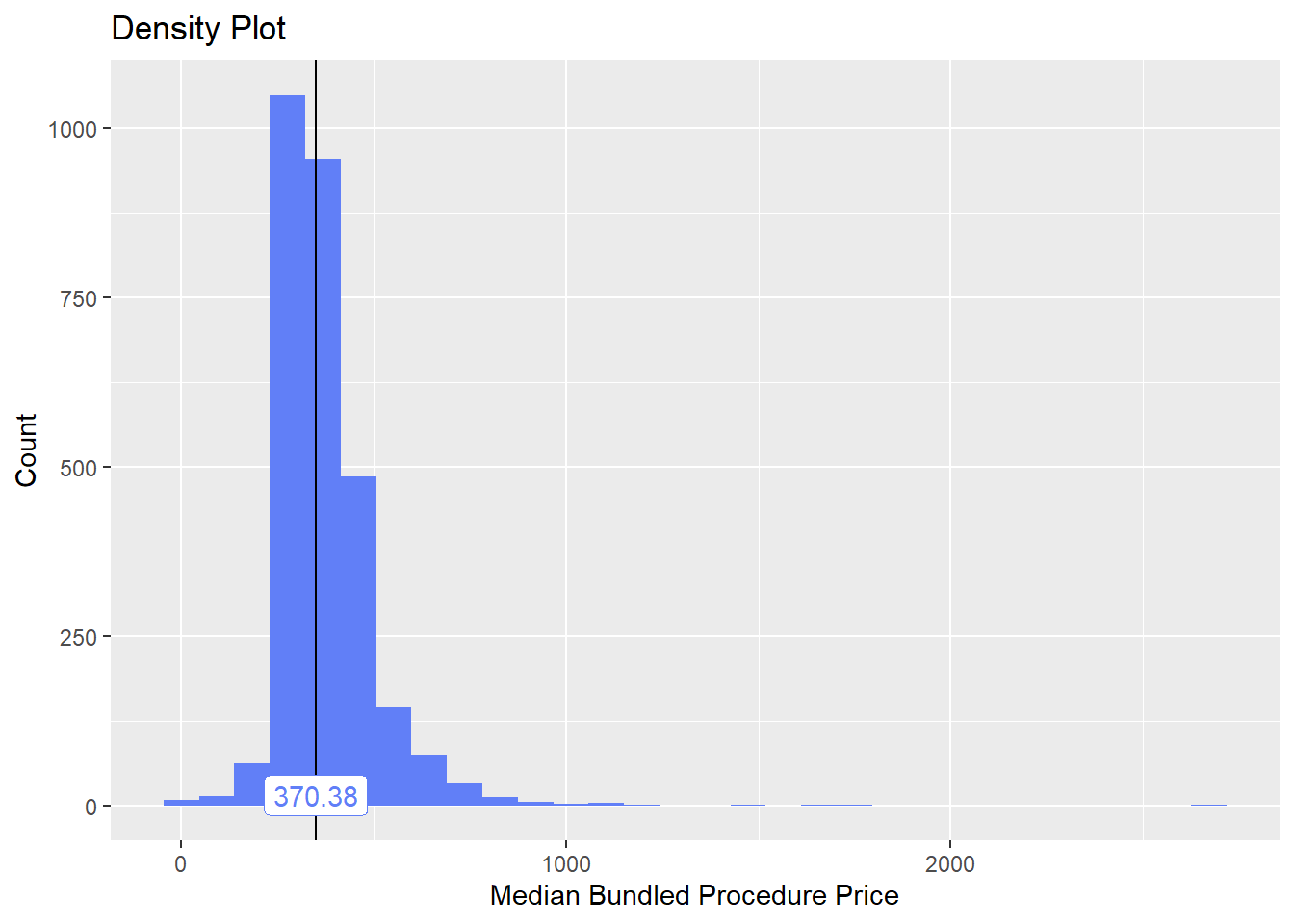
toe_nail_w_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 129 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_est 0.1
## 2 duration_mean 0.0866
## 3 duration_max 0.0866
## 4 medi_bun_t_inject 0.0645
## 5 radi_bun_t_ct 0.0625
## 6 radi_bun_t_without_dye 0.0625
## 7 medi_bun_t_manual 0.0619
## 8 path_bun_t_renal 0.0576
## 9 surg_bun_t_paravert 0.0567
## 10 medi_bun_t_hearing 0.0543
## # ... with 119 more rowstoe_nail_w_visit %>% get_tag_density_information("medi_bun_t_inject") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_inject"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 36.7## $dist_plots
##
## $stat_tables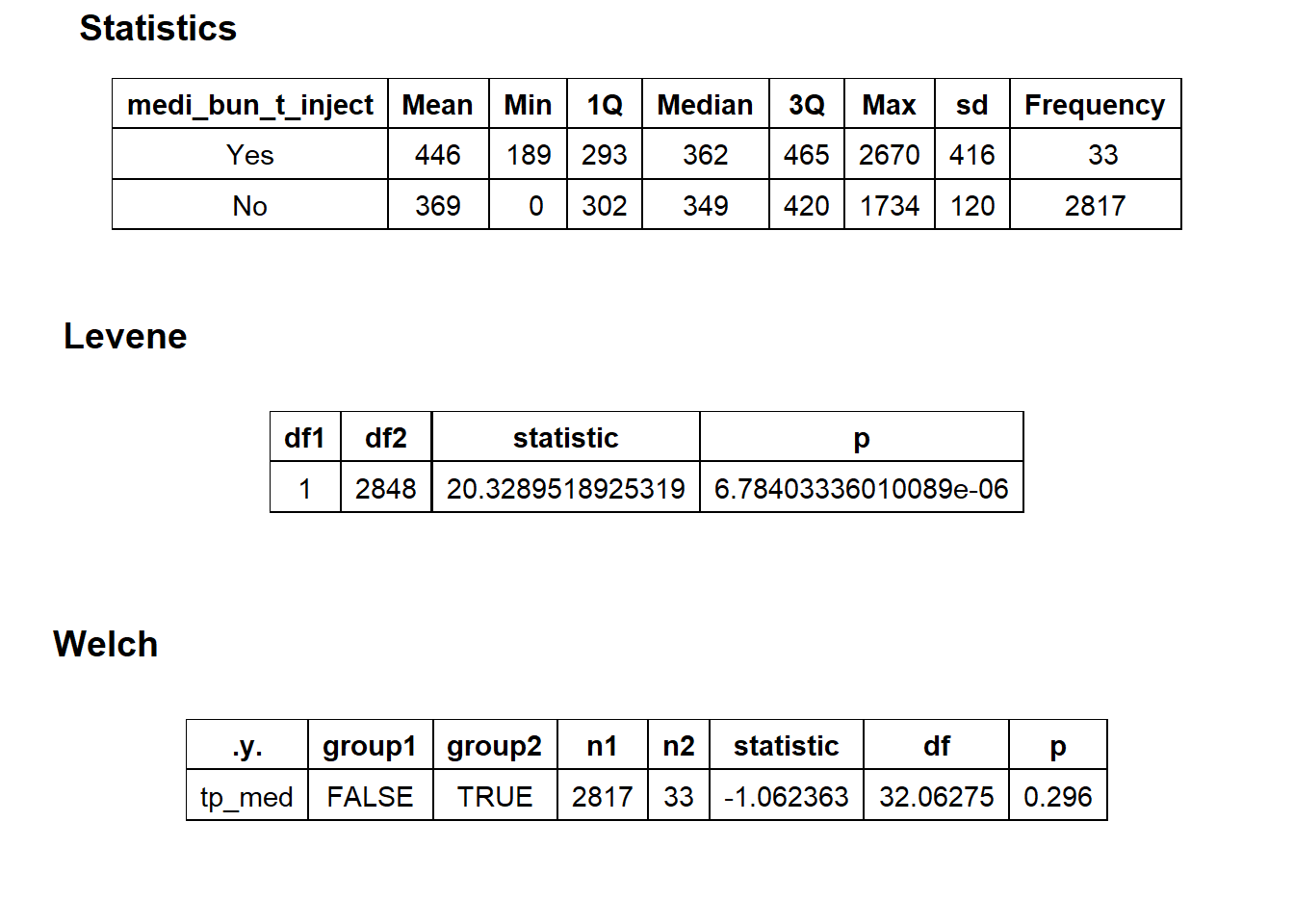
# only one observationtoe_nail_w_visit <- toe_nail_w_visit[medi_bun_t_inject==F]toe_nail_w_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.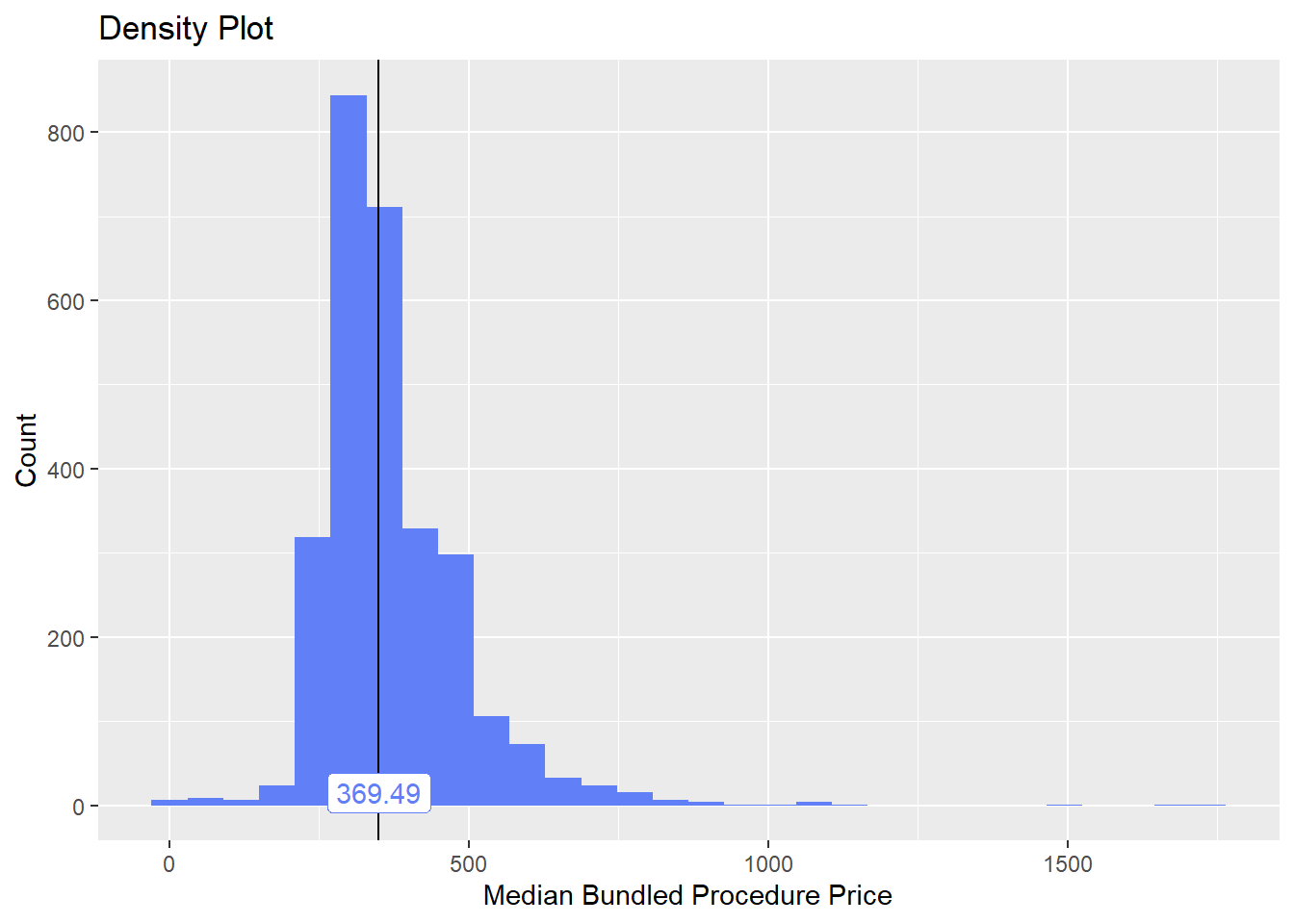
toe_nail_w_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 125 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_est 0.112
## 2 duration_mean 0.0927
## 3 duration_max 0.0927
## 4 radi_bun_t_ct 0.067
## 5 radi_bun_t_without_dye 0.067
## 6 medi_bun_t_manual 0.0663
## 7 path_bun_t_renal 0.0617
## 8 surg_bun_t_paravert 0.0607
## 9 medi_bun_t_boot 0.059
## 10 medi_bun_t_hearing 0.0582
## # ... with 115 more rowstoe_nail_w_visit %>% get_tag_density_information("radi_bun_t_ct") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ radi_bun_t_ct"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 16.7## $dist_plots
##
## $stat_tables
# only one observationtoe_nail_w_visit <- toe_nail_w_visit[radi_bun_t_ct==F]toe_nail_w_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail_w_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 123 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_est 0.113
## 2 duration_max 0.0929
## 3 duration_mean 0.0929
## 4 medi_bun_t_manual 0.0665
## 5 path_bun_t_renal 0.0619
## 6 surg_bun_t_paravert 0.0609
## 7 medi_bun_t_boot 0.0594
## 8 medi_bun_t_hearing 0.0584
## 9 medi_bun_t_medroxyprogesterone 0.0561
## 10 medi_bun_t_progesterone 0.0561
## # ... with 113 more rowstoe_nail_w_visit %>% get_tag_density_information("medi_bun_t_manual") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_manual"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 93.4## $dist_plots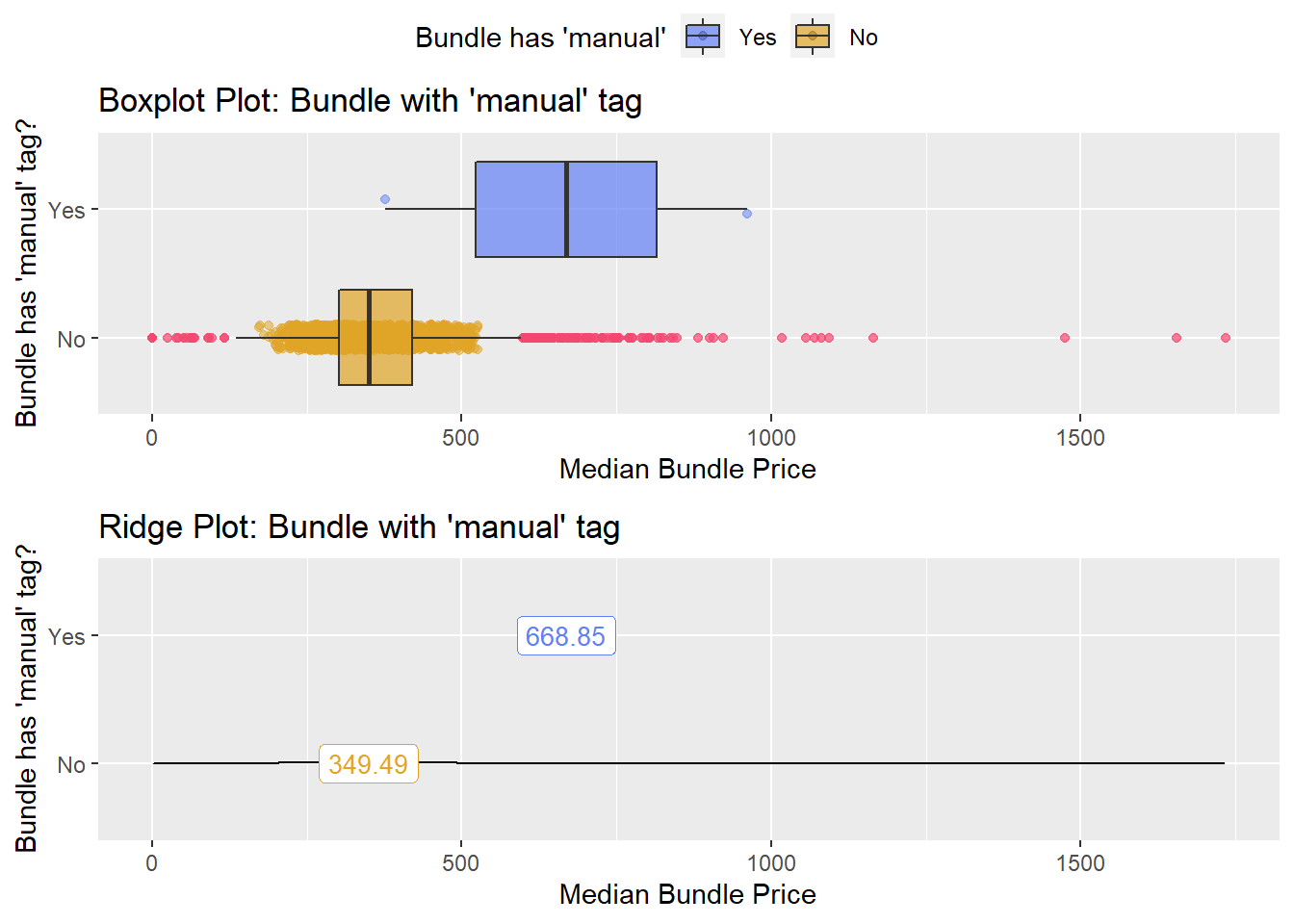
##
## $stat_tables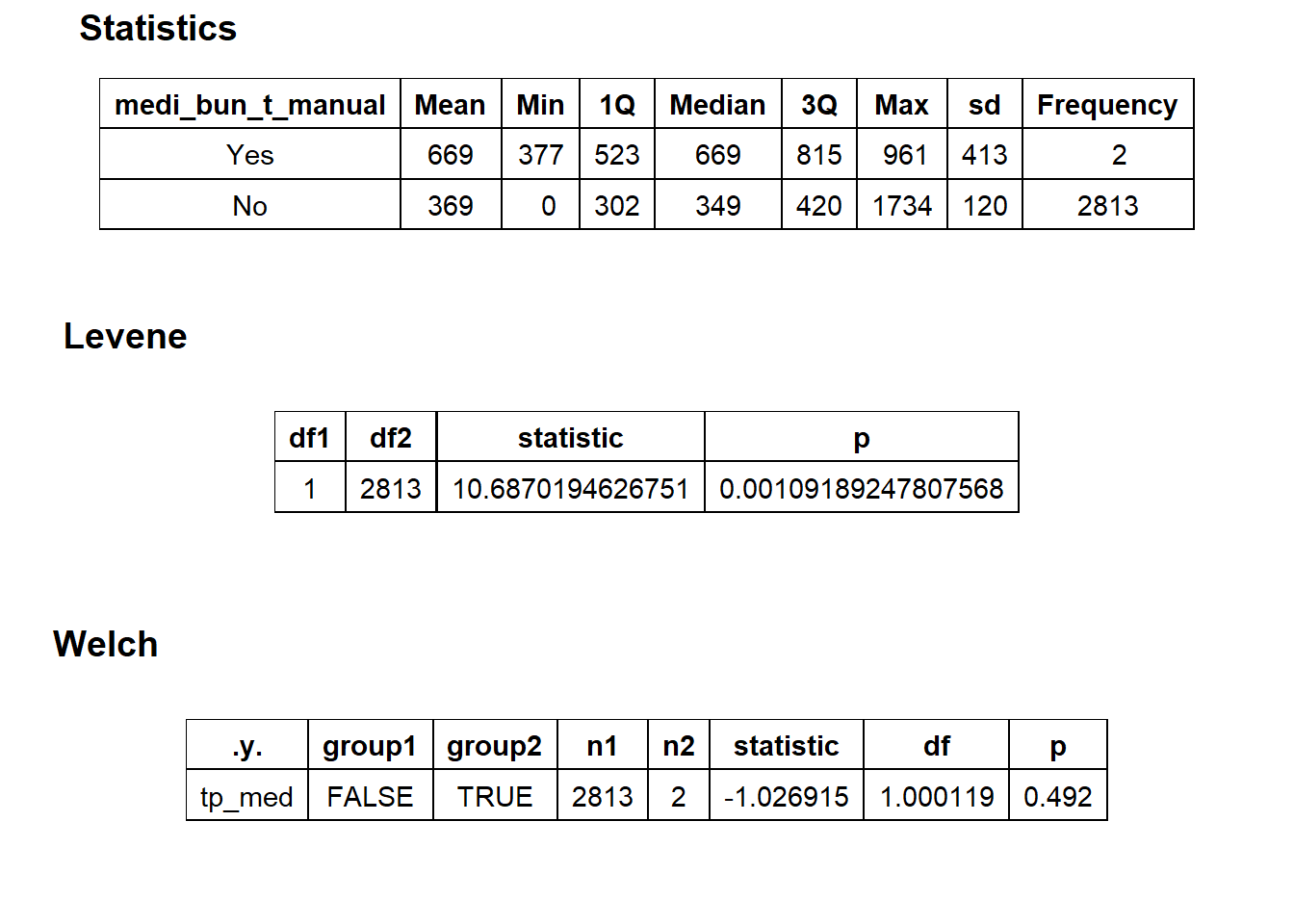
# only one observationtoe_nail_w_visit <- toe_nail_w_visit[medi_bun_t_manual==F]toe_nail_w_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.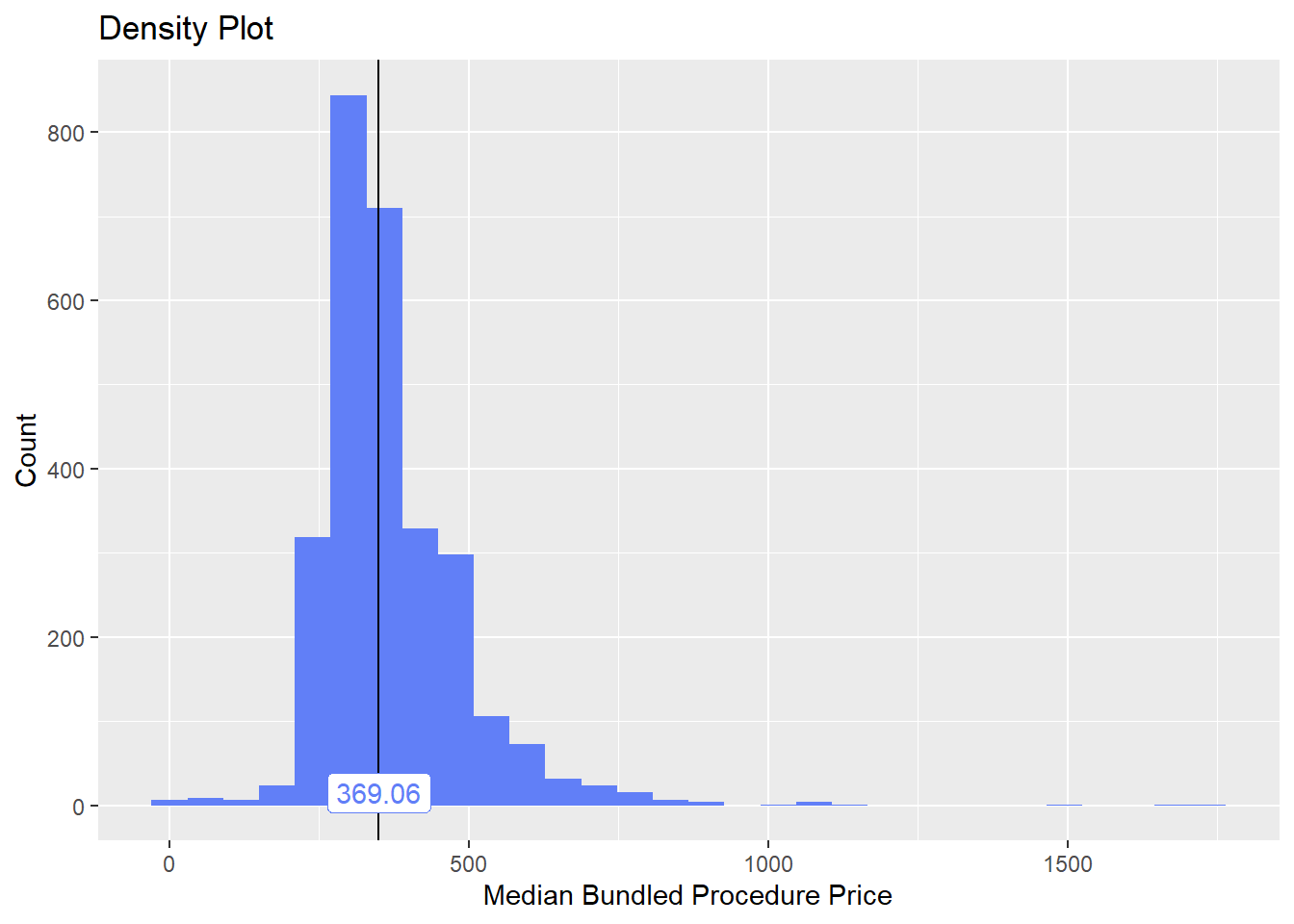
toe_nail_w_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 120 x 2
## name correlation
## <chr> <dbl>
## 1 faci_bun_t_est 0.116
## 2 path_bun_t_renal 0.0623
## 3 surg_bun_t_paravert 0.0612
## 4 medi_bun_t_boot 0.06
## 5 medi_bun_t_hearing 0.0587
## 6 medi_bun_t_medroxyprogesterone 0.0564
## 7 medi_bun_t_progesterone 0.0564
## 8 surg_bun_t_endoscopy 0.0528
## 9 surg_bun_t_nasal 0.0528
## 10 faci_bun_t_office 0.0508
## # ... with 110 more rowstoe_nail_w_visit %>% get_tag_density_information("path_bun_t_renal") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_renal"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 49.5## $dist_plots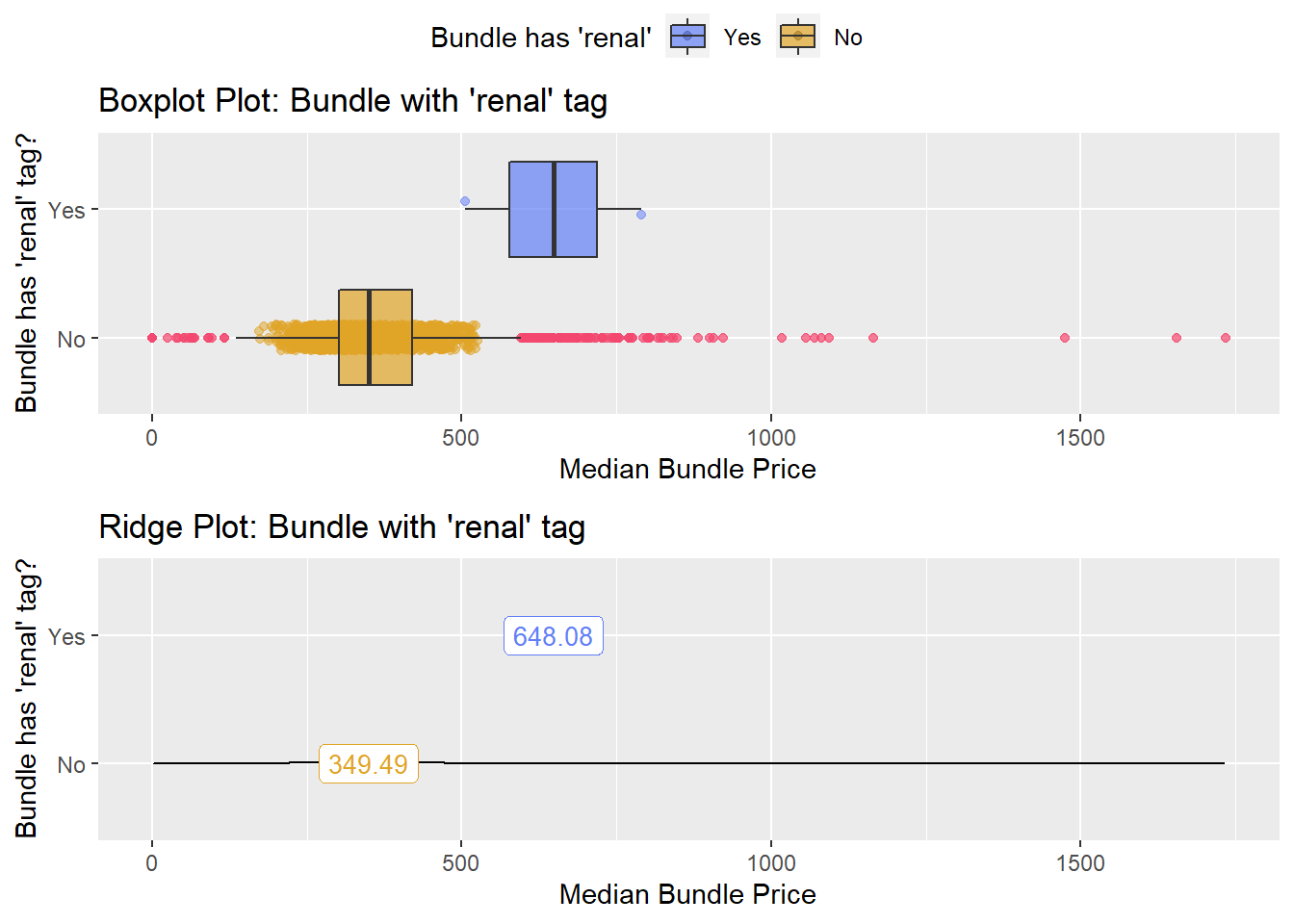
##
## $stat_tables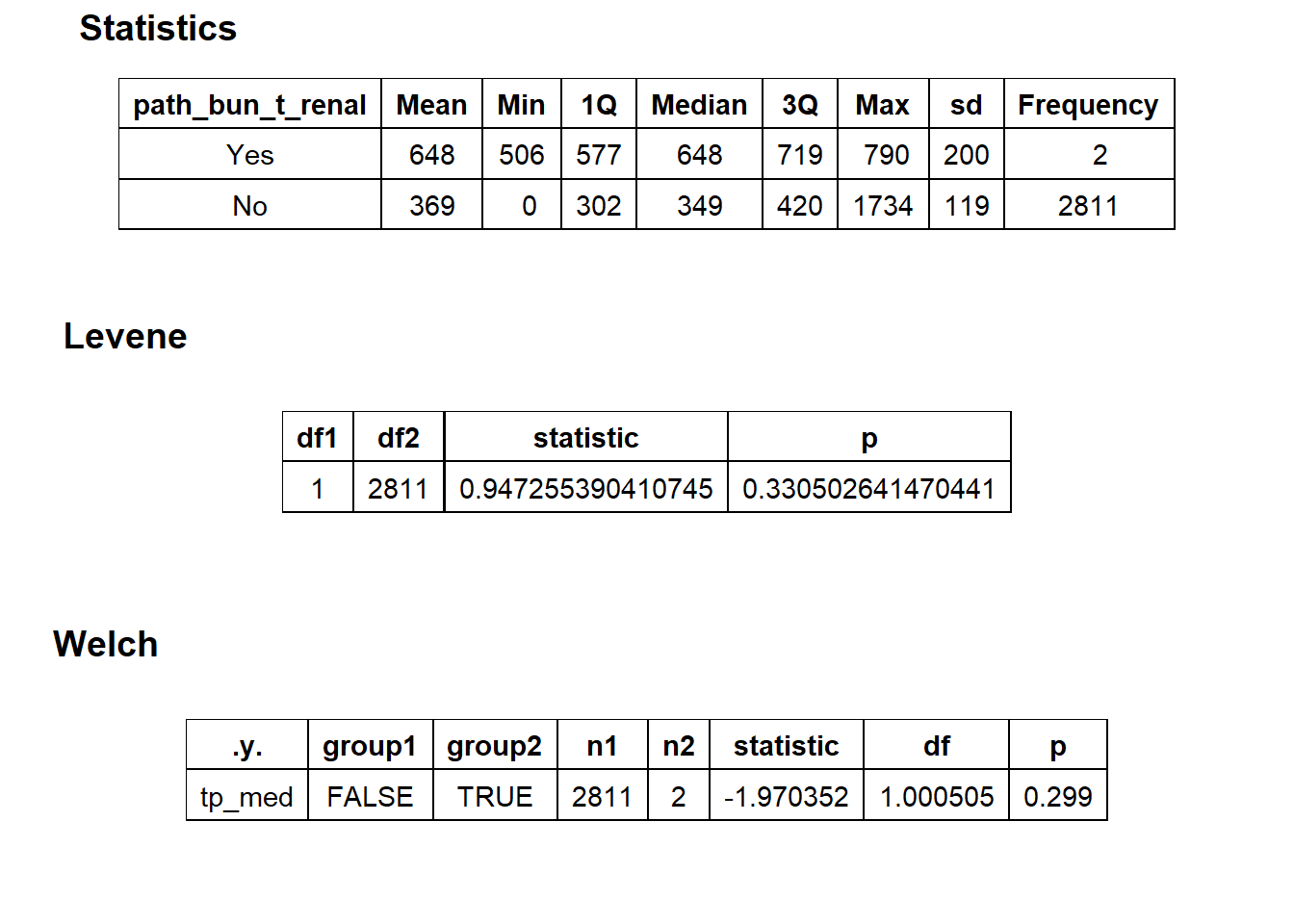
# only one observationtoe_nail_w_visit <- toe_nail_w_visit[path_bun_t_renal==F]toe_nail_w_visit <- toe_nail_w_visit[,`:=`(
surg_sp_name_clean = surg_sp_npi %>% map_chr(get_npi_standard_name),
surg_bp_name_clean = surg_bp_npi %>% map_chr(get_npi_standard_name),
medi_sp_name_clean = medi_sp_npi %>% map_chr(get_npi_standard_name),
medi_bp_name_clean = medi_bp_npi %>% map_chr(get_npi_standard_name),
radi_sp_name_clean = radi_sp_npi %>% map_chr(get_npi_standard_name),
radi_bp_name_clean = radi_bp_npi %>% map_chr(get_npi_standard_name),
path_sp_name_clean = path_sp_npi %>% map_chr(get_npi_standard_name),
path_bp_name_clean = path_bp_npi %>% map_chr(get_npi_standard_name),
anes_sp_name_clean = anes_sp_npi %>% map_chr(get_npi_standard_name),
anes_bp_name_clean = anes_bp_npi %>% map_chr(get_npi_standard_name),
faci_sp_name_clean = faci_sp_npi %>% map_chr(get_npi_standard_name),
faci_bp_name_clean = faci_bp_npi %>% map_chr(get_npi_standard_name)
)]499
toe_nail_w_visit_btbv5 <- toe_nail_w_visit %>%
btbv5(
osa_group_p=NA,
osa_class_p = "Podiatrist",
osa_specialization_p = NA
) %>%
.[,`:=`(procedure_type=23,
procedure_modifier="Standard",
ingest_date =Sys.Date())]## [1] "multiple meet class req"
## [1] "RYAN ELLSWORTH" "KATHRYN C DECKER"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"
## [1] "multiple meet class req"
## [1] "K2 FOOT AND ANKLE P (MURRAY)" "STANTON MILLER SMITH"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"
## [1] "multiple meet class req"
## [1] "DANIEL J HUFF" "ANDREW HARRIS BURGON"
## [1] "multiple meet class req"
## [1] "DANIEL J HUFF" "ANDREW HARRIS BURGON"
## [1] "multiple meet class req"
## [1] "K2 FOOT AND ANKLE P (MURRAY)" "STANTON MILLER SMITH"
## [1] "multiple meet class req"
## [1] "DAVID H WARBY DPM (LAYTON)" "DAVID H WARBY"
## [1] "multiple meet class req"
## [1] "CORAL DESERT FOOT ANKLE (ST GEORGE)"
## [2] "LANDON CAMERON"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"
## [1] "multiple meet class req"
## [1] "DANIEL J HUFF" "ANDREW HARRIS BURGON"
## [1] "multiple meet class req"
## [1] "D ALDEN YATES DPM (ST GEORGE)" "DON A YATES"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"
## [1] "multiple meet class req"
## [1] "DAVID JARAMILLO" "TODD JOSEPH JARAMILLO"
## [1] "multiple meet class req"
## [1] "SCOTT STEVEN SHELTON" "WADE A CHRISTIANSEN"
## [1] "multiple meet class req"
## [1] "JESSE RILEY DPM (PAYSON)" "JESSE ELLSWORTH RILEY"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"
## [1] "multiple meet class req"
## [1] "L CRAIG LARSEN DPM (MURRAY)" "CLARK C LARSEN"
## [1] "multiple meet class req"
## [1] "L CRAIG LARSEN DPM (MURRAY)" "CLARK C LARSEN"
## [1] "multiple meet class req"
## [1] "SCRODCO P (SOUTHWEST FOOT & ANKLE)" "RYAN THOMAS PETERSON"
## [1] "multiple meet class req"
## [1] "L CRAIG LARSEN DPM (MURRAY)" "CLARK C LARSEN"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"
## [1] "multiple meet class req"
## [1] "L CRAIG LARSEN DPM (MURRAY)" "CLARK C LARSEN"
## [1] "multiple meet class req"
## [1] "DAVID H WARBY DPM (LAYTON)" "DAVID H WARBY"
## [1] "multiple meet class req"
## [1] "CORAL DESERT FOOT ANKLE (ST GEORGE)"
## [2] "JEFFREY LEE STEWART"
## [1] "multiple meet class req"
## [1] "DAVID H WARBY DPM (LAYTON)" "DAVID H WARBY"
## [1] "multiple meet class req"
## [1] "DAVID H WARBY DPM (LAYTON)" "DAVID H WARBY"
## [1] "multiple meet class req"
## [1] "CORAL DESERT FOOT ANKLE (ST GEORGE)"
## [2] "LANDON CAMERON"
## [1] "multiple meet class req"
## [1] "DANIEL J HUFF" "ANDREW HARRIS BURGON"
## [1] "multiple meet class req"
## [1] "DAVID H WARBY DPM (LAYTON)" "DAVID H WARBY"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"
## [1] "multiple meet class req"
## [1] "D ALDEN YATES DPM (ST GEORGE)" "DON A YATES"
## [1] "multiple meet class req"
## [1] "L CRAIG LARSEN DPM (MURRAY)" "CLARK C LARSEN"
## [1] "multiple meet class req"
## [1] "DR BRIAN RICHMAN (LAYTON)" "BRIAN H RICHMAN"
## [1] "multiple meet class req"
## [1] "DAVID JARAMILLO" "TODD JOSEPH JARAMILLO"
## [1] "multiple meet class req"
## [1] "K2 FOOT AND ANKLE P (MURRAY)" "STANTON MILLER SMITH"
## [1] "multiple meet class req"
## [1] "DANIEL J HUFF" "ANDREW HARRIS BURGON"
## [1] "multiple meet class req"
## [1] "ANDREW H BURGON PODIATRY (SUMMIT FOOT AND ANKLE)"
## [2] "ANDREW HARRIS BURGON"
## [1] "multiple meet class req"
## [1] "RYAN ELLSWORTH" "KATHRYN C DECKER"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"
## [1] "multiple meet class req"
## [1] "SCRODCO P (SOUTHWEST FOOT & ANKLE)" "RYAN THOMAS PETERSON"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"
## [1] "multiple meet class req"
## [1] "CORAL DESERT FOOT ANKLE (ST GEORGE)"
## [2] "LANDON CAMERON"
## [1] "multiple meet class req"
## [1] "RYAN ELLSWORTH" "KATHRYN C DECKER"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"toe_nail_w_visit_btbv5$tp_med_med %>% summary()## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 1.0 311.6 352.3 382.9 417.4 1165.4toe_q1 <- toe_nail_w_visit_btbv5$tp_med_med %>% quantile(.25)
toe_q3 <- toe_nail_w_visit_btbv5$tp_med_med %>% quantile(.75)
toe_iqr <- toe_nail_w_visit_btbv5$tp_med_med %>% IQR()toe_nail_w_visit_btbv5_final <- toe_nail_w_visit_btbv5[tp_med_med >toe_q1 -1.5*toe_iqr & tp_med_med < toe_q3 + 1.5*toe_iqr ]toe_nail_w_visit_btbv5$tp_med_med %>% summary()## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 1.0 311.6 352.3 382.9 417.4 1165.4toe_nail_w_visit_btbv5_final$tp_med_med %>% summary()## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 196.6 310.8 350.0 361.7 396.5 575.0bq_table_upload(x=procedure_dev_table, values= toe_nail_w_visit_btbv5_final, create_disposition='CREATE_IF_NEEDED', write_disposition='WRITE_APPEND')##toe_nail_n_visit
toe_nail_n_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.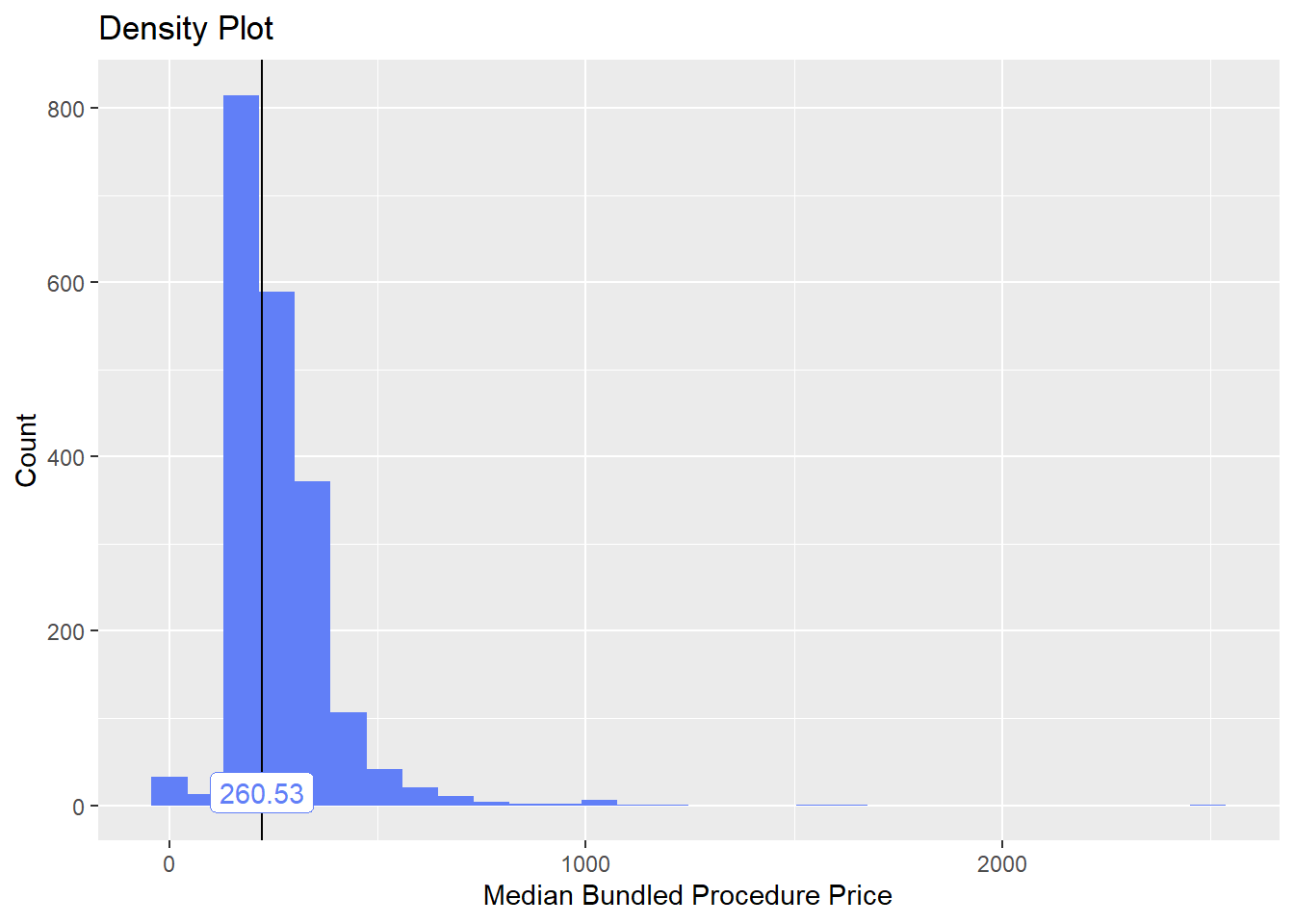
toe_nail_n_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 98 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_dev 0.157
## 2 surg_bun_t_device 0.157
## 3 path_bun_t_dna 0.157
## 4 path_bun_t_gonorr 0.157
## 5 path_bun_t_pregnancy 0.157
## 6 path_bun_t_urine 0.157
## 7 medi_bun_t_psych 0.128
## 8 surg_bun_t_insert 0.108
## 9 surg_bun_t_incision 0.0951
## 10 surg_bun_t_toe 0.0951
## # ... with 88 more rows# toe_nail_n_visit %>% get_tag_density_information("surg_bun_t_dev") %>% print()
# only one observationtoe_nail_n_visit <- toe_nail_n_visit[surg_bun_t_dev==F]toe_nail_n_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail_n_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 92 x 2
## name correlation
## <chr> <dbl>
## 1 medi_bun_t_psych 0.130
## 2 surg_bun_t_incision 0.0965
## 3 surg_bun_t_toe 0.0965
## 4 medi_bun_t_boot 0.0959
## 5 surg_bun_t_endo 0.0628
## 6 surg_bun_t_tendon 0.0628
## 7 surg_bun_t_shave 0.0545
## 8 surg_bun_t_plate 0.052
## 9 surg_bun_t_interlaminar 0.0519
## 10 surg_bun_t_lamina 0.0519
## # ... with 82 more rows# toe_nail_n_visit %>% get_tag_density_information("medi_bun_t_psych") %>% print()
# only one oberservationtoe_nail_n_visit <- toe_nail_n_visit[medi_bun_t_psych==F]toe_nail_n_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail_n_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 91 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_incision 0.0974
## 2 surg_bun_t_toe 0.0974
## 3 medi_bun_t_boot 0.097
## 4 surg_bun_t_endo 0.0635
## 5 surg_bun_t_tendon 0.0635
## 6 surg_bun_t_shave 0.055
## 7 surg_bun_t_plate 0.0527
## 8 surg_bun_t_interlaminar 0.0524
## 9 surg_bun_t_lamina 0.0524
## 10 surg_bun_t_njx 0.0524
## # ... with 81 more rowstoe_nail_n_visit %>% get_tag_density_information("surg_bun_t_endo") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_endo"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 46.6## $dist_plots
##
## $stat_tables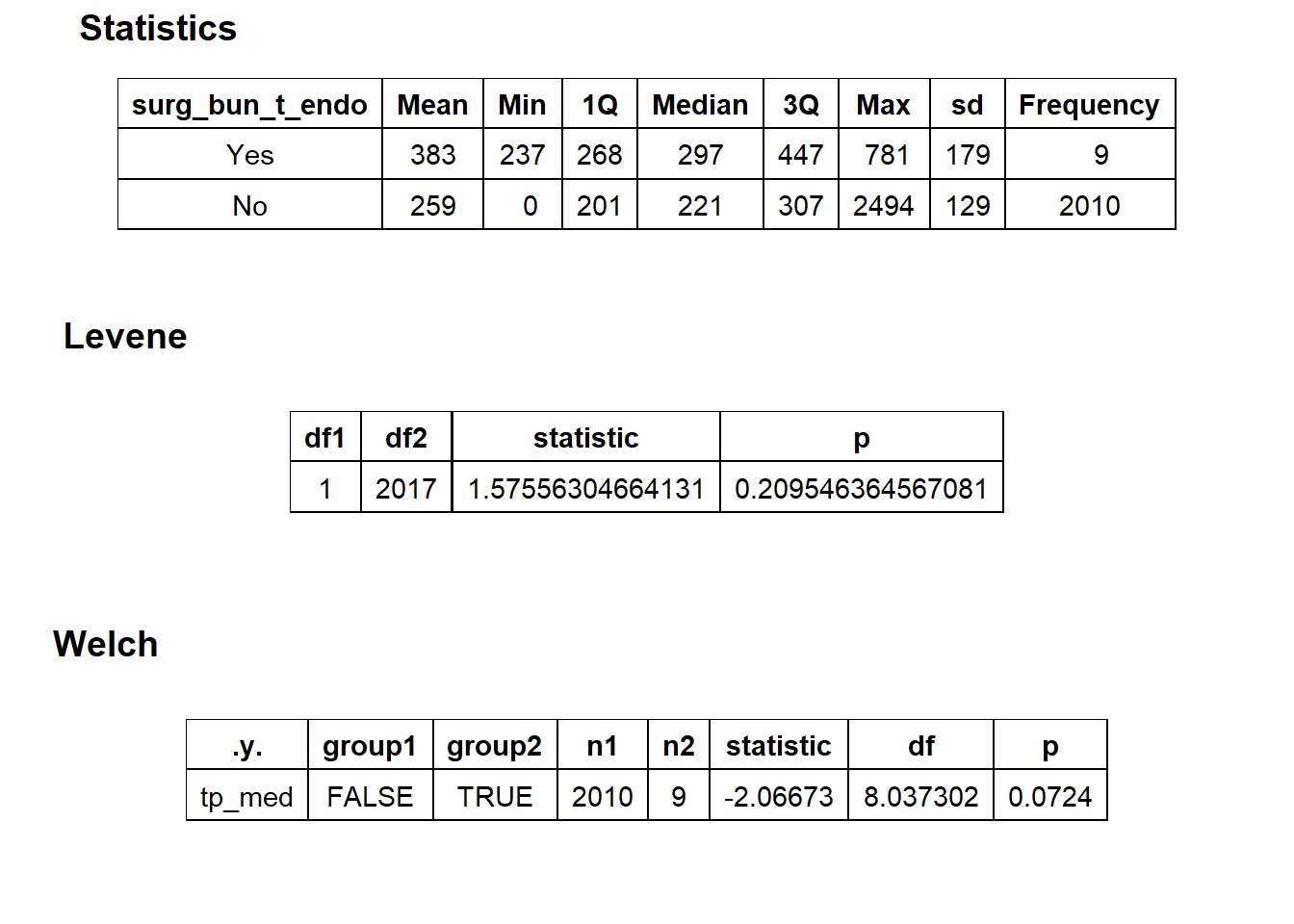
toe_nail_n_visit <- toe_nail_n_visit[surg_bun_t_endo==F]toe_nail_n_visit %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail_n_visit %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 84 x 2
## name correlation
## <chr> <dbl>
## 1 medi_bun_t_boot 0.098
## 2 surg_bun_t_shave 0.0554
## 3 surg_bun_t_plate 0.0533
## 4 surg_bun_t_interlaminar 0.0528
## 5 surg_bun_t_lamina 0.0528
## 6 surg_bun_t_njx 0.0528
## 7 radi_bun_t_mammo 0.0483
## 8 surg_bun_t_b9 0.0471
## 9 surg_bun_t_exc 0.0465
## 10 surg_bun_t_other 0.046
## # ... with 74 more rowscols_to_check <- toe_nail_n_visit %>% select_if(is.logical)
tag_frequency <- apply(cols_to_check, 2, function(i) sum(i > 0)) %>% enframe() %>% as.data.table() %>% .[,.(tag=name, freq=value)]
outlier_tag_list <- tag_frequency[freq < 2] %>% .[["tag"]]toe_nail_n_visit_clean <- remove_bun_proc_with_rare_tags(outlier_tag_list=outlier_tag_list, df=toe_nail_n_visit)## [1] "Start of rare tag removal"
## [1] "nrow:2010 , ncol:366"
## [1] "nrow:2010"
## [1] "removing: surg_bun_t_abscess"
## [1] 2009
## [1] "removing: surg_bun_t_amput"
## [1] 2009
## [1] "removing: surg_bun_t_ankle"
## [1] 2009
## [1] "removing: surg_bun_t_art"
## [1] 2009
## [1] "removing: surg_bun_t_arthro"
## [1] 2009
## [1] "removing: surg_bun_t_arthroscopy"
## [1] 2009
## [1] "removing: surg_bun_t_biopsy"
## [1] 2009
## [1] "removing: surg_bun_t_body"
## [1] 2009
## [1] "removing: surg_bun_t_bone"
## [1] 2009
## [1] "removing: surg_bun_t_compression"
## [1] 2009
## [1] "removing: surg_bun_t_correct"
## [1] 2009
## [1] "removing: surg_bun_t_cyst"
## [1] 2009
## [1] "removing: surg_bun_t_debrid"
## [1] 2009
## [1] "removing: surg_bun_t_debride"
## [1] 2009
## [1] "removing: surg_bun_t_decompression"
## [1] 2009
## [1] "removing: surg_bun_t_deformity"
## [1] 2009
## [1] "removing: surg_bun_t_dev"
## [1] 2009
## [1] "removing: surg_bun_t_device"
## [1] 2009
## [1] "removing: surg_bun_t_diagnostic"
## [1] 2009
## [1] "removing: surg_bun_t_dislocat"
## [1] 2009
## [1] "removing: surg_bun_t_drug"
## [1] 2009
## [1] "removing: surg_bun_t_duct"
## [1] 2009
## [1] "removing: surg_bun_t_dura"
## [1] 2009
## [1] "removing: surg_bun_t_eardrum"
## [1] 2008
## [1] "removing: surg_bun_t_egd"
## [1] 2008
## [1] "removing: surg_bun_t_endo"
## [1] 2008
## [1] "removing: surg_bun_t_endoscopy"
## [1] 2008
## [1] "removing: surg_bun_t_excision"
## [1] 2008
## [1] "removing: surg_bun_t_eye"
## [1] 2008
## [1] "removing: surg_bun_t_face"
## [1] 2007
## [1] "removing: surg_bun_t_fascia"
## [1] 2007
## [1] "removing: surg_bun_t_finger"
## [1] 2007
## [1] "removing: surg_bun_t_foot"
## [1] 2006
## [1] "removing: surg_bun_t_foreign"
## [1] 2006
## [1] "removing: surg_bun_t_fracture"
## [1] 2006
## [1] "removing: surg_bun_t_fusion"
## [1] 2006
## [1] "removing: surg_bun_t_graft"
## [1] 2006
## [1] "removing: surg_bun_t_hallux"
## [1] 2006
## [1] "removing: surg_bun_t_hand"
## [1] 2006
## [1] "removing: surg_bun_t_implant"
## [1] 2006
## [1] "removing: surg_bun_t_incision"
## [1] 2006
## [1] "removing: surg_bun_t_insert"
## [1] 2006
## [1] "removing: surg_bun_t_interlaminar"
## [1] 2005
## [1] "removing: surg_bun_t_knee"
## [1] 2005
## [1] "removing: surg_bun_t_lamina"
## [1] 2005
## [1] "removing: surg_bun_t_leg"
## [1] 2005
## [1] "removing: surg_bun_t_ligament"
## [1] 2005
## [1] "removing: surg_bun_t_lower"
## [1] 2005
## [1] "removing: surg_bun_t_nasal"
## [1] 2005
## [1] "removing: surg_bun_t_nerve"
## [1] 2005
## [1] "removing: surg_bun_t_njx"
## [1] 2005
## [1] "removing: surg_bun_t_open"
## [1] 2005
## [1] "removing: surg_bun_t_opening"
## [1] 2005
## [1] "removing: surg_bun_t_paravert"
## [1] 2005
## [1] "removing: surg_bun_t_part"
## [1] 2005
## [1] "removing: surg_bun_t_partial"
## [1] 2005
## [1] "removing: surg_bun_t_release"
## [1] 2005
## [1] "removing: surg_bun_t_repair"
## [1] 2005
## [1] "removing: surg_bun_t_resect"
## [1] 2005
## [1] "removing: surg_bun_t_revise"
## [1] 2005
## [1] "removing: surg_bun_t_revision"
## [1] 2005
## [1] "removing: surg_bun_t_shave"
## [1] 2004
## [1] "removing: surg_bun_t_sheath"
## [1] 2004
## [1] "removing: surg_bun_t_sperm"
## [1] 2004
## [1] "removing: surg_bun_t_sub"
## [1] 2004
## [1] "removing: surg_bun_t_tendon"
## [1] 2004
## [1] "removing: surg_bun_t_thumb"
## [1] 2004
## [1] "removing: surg_bun_t_tip"
## [1] 2004
## [1] "removing: surg_bun_t_tiss"
## [1] 2004
## [1] "removing: surg_bun_t_toe"
## [1] 2004
## [1] "removing: surg_bun_t_treat"
## [1] 2004
## [1] "removing: surg_bun_t_tum"
## [1] 2004
## [1] "removing: surg_bun_t_unit"
## [1] 2004
## [1] "removing: surg_bun_t_valgus"
## [1] 2004
## [1] "removing: surg_bun_t_wrist"
## [1] 2004
## [1] "removing: medi_bun_t_add-on"
## [1] 2004
## [1] "removing: medi_bun_t_analy"
## [1] 2004
## [1] "removing: medi_bun_t_choline"
## [1] 2004
## [1] "removing: medi_bun_t_clinic"
## [1] 2003
## [1] "removing: medi_bun_t_depression"
## [1] 2003
## [1] "removing: medi_bun_t_establish"
## [1] 2003
## [1] "removing: medi_bun_t_evaluat"
## [1] 2003
## [1] "removing: medi_bun_t_exercise"
## [1] 2003
## [1] "removing: medi_bun_t_hearing"
## [1] 2003
## [1] "removing: medi_bun_t_hpv"
## [1] 2003
## [1] "removing: medi_bun_t_hydrate"
## [1] 2003
## [1] "removing: medi_bun_t_immune"
## [1] 2003
## [1] "removing: medi_bun_t_laiv4"
## [1] 2003
## [1] "removing: medi_bun_t_medroxyprogesterone"
## [1] 2003
## [1] "removing: medi_bun_t_methylprednisolone"
## [1] 2003
## [1] "removing: medi_bun_t_oral"
## [1] 2003
## [1] "removing: medi_bun_t_outpt"
## [1] 2003
## [1] "removing: medi_bun_t_prednisolone"
## [1] 2003
## [1] "removing: medi_bun_t_progesterone"
## [1] 2003
## [1] "removing: medi_bun_t_psych"
## [1] 2003
## [1] "removing: medi_bun_t_reeducation"
## [1] 2003
## [1] "removing: medi_bun_t_services"
## [1] 2003
## [1] "removing: medi_bun_t_solution"
## [1] 2003
## [1] "removing: medi_bun_t_sound"
## [1] 2003
## [1] "removing: medi_bun_t_stud"
## [1] 2003
## [1] "removing: medi_bun_t_test"
## [1] 2002
## [1] "removing: medi_bun_t_testosterone"
## [1] 2002
## [1] "removing: medi_bun_t_therapeutic"
## [1] 2001
## [1] "removing: medi_bun_t_tracing"
## [1] 2001
## [1] "removing: medi_bun_t_traction"
## [1] 2001
## [1] "removing: medi_bun_t_ultrasound"
## [1] 2001
## [1] "removing: medi_bun_t_whirlpool"
## [1] 2001
## [1] "removing: medi_bun_t_wound"
## [1] 2001
## [1] "removing: medi_bun_t_hepa"
## [1] 2001
## [1] "removing: medi_bun_t_psytx"
## [1] 2001
## [1] "removing: medi_bun_t_pcv13"
## [1] 2000
## [1] "removing: medi_bun_t_eye_exam"
## [1] 2000
## [1] "removing: medi_bun_t_menacwy"
## [1] 2000
## [1] "removing: radi_bun_t_ankle"
## [1] 2000
## [1] "removing: radi_bun_t_bone"
## [1] 2000
## [1] "removing: radi_bun_t_breast"
## [1] 2000
## [1] "removing: radi_bun_t_chest"
## [1] 2000
## [1] "removing: radi_bun_t_ct"
## [1] 2000
## [1] "removing: radi_bun_t_echo"
## [1] 2000
## [1] "removing: radi_bun_t_exam"
## [1] 2000
## [1] "removing: radi_bun_t_finger"
## [1] 2000
## [1] "removing: radi_bun_t_fluoroscopy"
## [1] 2000
## [1] "removing: radi_bun_t_foot"
## [1] 2000
## [1] "removing: radi_bun_t_guide"
## [1] 2000
## [1] "removing: radi_bun_t_hand"
## [1] 2000
## [1] "removing: radi_bun_t_head"
## [1] 2000
## [1] "removing: radi_bun_t_joint"
## [1] 2000
## [1] "removing: radi_bun_t_lumbar"
## [1] 2000
## [1] "removing: radi_bun_t_mammo"
## [1] 1999
## [1] "removing: radi_bun_t_mri"
## [1] 1999
## [1] "removing: radi_bun_t_neck"
## [1] 1999
## [1] "removing: radi_bun_t_pelvi"
## [1] 1999
## [1] "removing: radi_bun_t_shoulder"
## [1] 1999
## [1] "removing: radi_bun_t_spine"
## [1] 1999
## [1] "removing: radi_bun_t_thorac"
## [1] 1999
## [1] "removing: radi_bun_t_toe"
## [1] 1999
## [1] "removing: radi_bun_t_view"
## [1] 1999
## [1] "removing: radi_bun_t_wrist"
## [1] 1999
## [1] "removing: radi_bun_t_without_dye"
## [1] 1999
## [1] "removing: radi_bun_t_x-ray"
## [1] 1999
## [1] "removing: path_bun_t_acid"
## [1] 1998
## [1] "removing: path_bun_t_agent"
## [1] 1998
## [1] "removing: path_bun_t_anti"
## [1] 1998
## [1] "removing: path_bun_t_antibod"
## [1] 1998
## [1] "removing: path_bun_t_bact"
## [1] 1998
## [1] "removing: path_bun_t_bacteria"
## [1] 1998
## [1] "removing: path_bun_t_blood"
## [1] 1998
## [1] "removing: path_bun_t_colon"
## [1] 1998
## [1] "removing: path_bun_t_count"
## [1] 1997
## [1] "removing: path_bun_t_cpk"
## [1] 1997
## [1] "removing: path_bun_t_creatinine"
## [1] 1997
## [1] "removing: path_bun_t_cytopath"
## [1] 1997
## [1] "removing: path_bun_t_detect"
## [1] 1997
## [1] "removing: path_bun_t_dna"
## [1] 1997
## [1] "removing: path_bun_t_drug"
## [1] 1997
## [1] "removing: path_bun_t_enzyme"
## [1] 1996
## [1] "removing: path_bun_t_factor"
## [1] 1996
## [1] "removing: path_bun_t_fecal"
## [1] 1996
## [1] "removing: path_bun_t_ferritin"
## [1] 1996
## [1] "removing: path_bun_t_fluid"
## [1] 1996
## [1] "removing: path_bun_t_free"
## [1] 1996
## [1] "removing: path_bun_t_function"
## [1] 1996
## [1] "removing: path_bun_t_gene"
## [1] 1995
## [1] "removing: path_bun_t_general"
## [1] 1995
## [1] "removing: path_bun_t_giardia"
## [1] 1995
## [1] "removing: path_bun_t_glucose"
## [1] 1995
## [1] "removing: path_bun_t_gonorr"
## [1] 1995
## [1] "removing: path_bun_t_health"
## [1] 1995
## [1] "removing: path_bun_t_hep"
## [1] 1995
## [1] "removing: path_bun_t_hepati"
## [1] 1995
## [1] "removing: path_bun_t_hiv"
## [1] 1994
## [1] "removing: path_bun_t_hormone"
## [1] 1993
## [1] "removing: path_bun_t_immunohisto"
## [1] 1993
## [1] "removing: path_bun_t_iron"
## [1] 1993
## [1] "removing: path_bun_t_lipid"
## [1] 1993
## [1] "removing: path_bun_t_lipoprotein"
## [1] 1993
## [1] "removing: path_bun_t_magnesium"
## [1] 1993
## [1] "removing: path_bun_t_patho"
## [1] 1993
## [1] "removing: path_bun_t_pathologist"
## [1] 1993
## [1] "removing: path_bun_t_phosphorus"
## [1] 1993
## [1] "removing: path_bun_t_potassium"
## [1] 1993
## [1] "removing: path_bun_t_pregnancy"
## [1] 1993
## [1] "removing: path_bun_t_progesterone"
## [1] 1993
## [1] "removing: path_bun_t_protein"
## [1] 1993
## [1] "removing: path_bun_t_psa"
## [1] 1993
## [1] "removing: path_bun_t_quantitative"
## [1] 1993
## [1] "removing: path_bun_t_renal"
## [1] 1993
## [1] "removing: path_bun_t_sex"
## [1] 1993
## [1] "removing: path_bun_t_smear"
## [1] 1993
## [1] "removing: path_bun_t_strep"
## [1] 1992
## [1] "removing: path_bun_t_testosterone"
## [1] 1992
## [1] "removing: path_bun_t_thyroid"
## [1] 1992
## [1] "removing: path_bun_t_time"
## [1] 1992
## [1] "removing: path_bun_t_tissue"
## [1] 1992
## [1] "removing: path_bun_t_urinalysis"
## [1] 1992
## [1] "removing: path_bun_t_urine"
## [1] 1992
## [1] "removing: path_bun_t_virus"
## [1] 1992
## [1] "removing: path_bun_t_vitamin"
## [1] 1992
## [1] "removing: path_bun_t_without_scope"
## [1] 1992
## [1] "removing: anes_bun_t_anes"
## [1] 1992
## [1] "removing: anes_bun_t_anesth"
## [1] 1992
## [1] "removing: anes_bun_t_bone"
## [1] 1992
## [1] "removing: anes_bun_t_knee"
## [1] 1992
## [1] "removing: anes_bun_t_leg"
## [1] 1992
## [1] "removing: anes_bun_t_lower"
## [1] 1992
## [1] "removing: anes_bun_t_nose"
## [1] 1992
## [1] "removing: anes_bun_t_sinus"
## [1] 1992
## [1] "removing: anes_bun_t_surg"
## [1] 1992
## [1] "removing: faci_bun_t_dept"
## [1] 1992
## [1] "removing: faci_bun_t_emergency"
## [1] 1992
## [1] "removing: faci_bun_t_est"
## [1] 1992
## [1] "removing: faci_bun_t_follow-up"
## [1] 1992
## [1] "removing: faci_bun_t_handling"
## [1] 1991
## [1] "removing: faci_bun_t_lab"
## [1] 1991
## [1] "removing: faci_bun_t_office"
## [1] 1991
## [1] "removing: faci_bun_t_pat"
## [1] 1991
## [1] "removing: faci_bun_t_postop"
## [1] 1991
## [1] "removing: faci_bun_t_specimen"
## [1] 1991
## [1] "removing: faci_bun_t_visit"
## [1] 1991toe_nail_n_visit_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.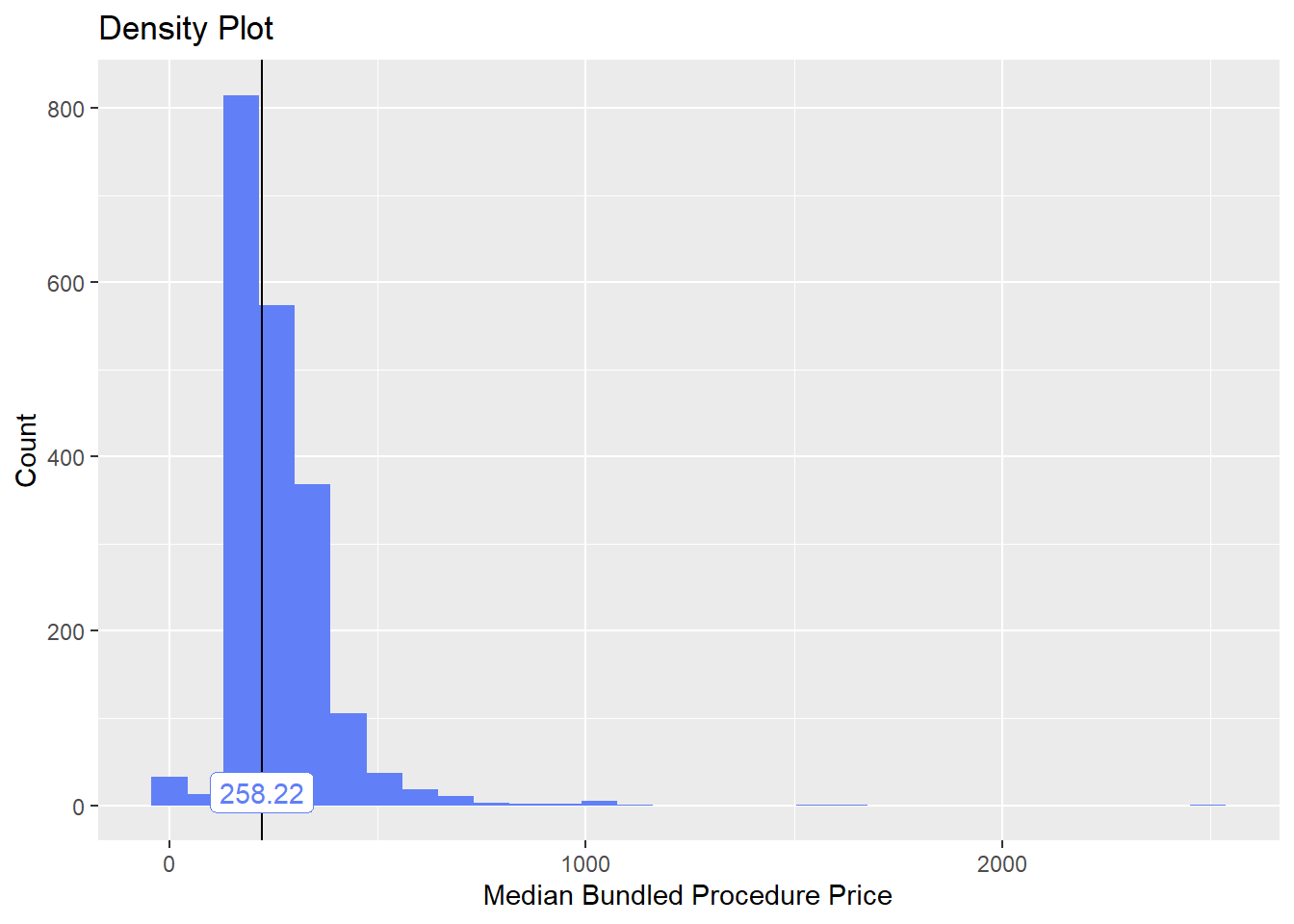
toe_nail_n_visit_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 50 x 2
## name correlation
## <chr> <dbl>
## 1 medi_bun_t_boot 0.0995
## 2 surg_bun_t_other 0.0466
## 3 surg_bun_t_plate 0.0403
## 4 medi_bun_t_visit 0.0398
## 5 medi_bun_t_immunization 0.0368
## 6 medi_bun_t_immun 0.0356
## 7 surg_bun_t_b9 0.0342
## 8 path_bun_t_hemoglobin 0.0331
## 9 medi_bun_t_vacc 0.0301
## 10 medi_bun_t_iiv 0.0276
## # ... with 40 more rowstoe_nail_n_visit_clean %>% get_tag_density_information("medi_bun_t_boot") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_boot"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 46.2## $dist_plots
##
## $stat_tables
toe_nail_n_visit_clean <- toe_nail_n_visit_clean[medi_bun_t_boot == F]toe_nail_n_visit_clean %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
toe_nail_n_visit_clean %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 49 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_other 0.0477
## 2 surg_bun_t_plate 0.0416
## 3 medi_bun_t_visit 0.04
## 4 medi_bun_t_immunization 0.0385
## 5 medi_bun_t_immun 0.0373
## 6 surg_bun_t_b9 0.0355
## 7 path_bun_t_hemoglobin 0.0338
## 8 medi_bun_t_vacc 0.0318
## 9 medi_bun_t_iiv 0.029
## 10 surg_bun_t_lesion 0.0261
## # ... with 39 more rowstoe_nail_n_visit_clean <- toe_nail_n_visit_clean[,`:=`(
surg_sp_name_clean = surg_sp_npi %>% map_chr(get_npi_standard_name),
surg_bp_name_clean = surg_bp_npi %>% map_chr(get_npi_standard_name),
medi_sp_name_clean = medi_sp_npi %>% map_chr(get_npi_standard_name),
medi_bp_name_clean = medi_bp_npi %>% map_chr(get_npi_standard_name),
radi_sp_name_clean = radi_sp_npi %>% map_chr(get_npi_standard_name),
radi_bp_name_clean = radi_bp_npi %>% map_chr(get_npi_standard_name),
path_sp_name_clean = path_sp_npi %>% map_chr(get_npi_standard_name),
path_bp_name_clean = path_bp_npi %>% map_chr(get_npi_standard_name),
anes_sp_name_clean = anes_sp_npi %>% map_chr(get_npi_standard_name),
anes_bp_name_clean = anes_bp_npi %>% map_chr(get_npi_standard_name),
faci_sp_name_clean = faci_sp_npi %>% map_chr(get_npi_standard_name),
faci_bp_name_clean = faci_bp_npi %>% map_chr(get_npi_standard_name)
)]499
toe_nail_n_visit_clean_btbv5 <- toe_nail_n_visit_clean %>%
btbv5(
osa_group_p=NA,
osa_class_p = "Podiatrist",
osa_specialization_p = NA
) %>%
.[,`:=`(procedure_type=23,
procedure_modifier="No Visit",
ingest_date =Sys.Date())]## [1] "multiple meet class req"
## [1] "DR BRIAN RICHMAN (LAYTON)" "BRIAN H RICHMAN"
## [1] "multiple meet class req"
## [1] "SCRODCO P (SOUTHWEST FOOT & ANKLE)" "DAVID T MAGNESEN"
## [1] "multiple meet class req"
## [1] "DR BRIAN RICHMAN (LAYTON)" "BRIAN H RICHMAN"
## [1] "multiple meet class req"
## [1] "CORAL DESERT FOOT ANKLE (ST GEORGE)"
## [2] "LANDON CAMERON"
## [1] "multiple meet class req"
## [1] "SCRODCO P (SOUTHWEST FOOT & ANKLE)" "RYAN THOMAS PETERSON"
## [1] "multiple meet class req"
## [1] "CORAL DESERT FOOT ANKLE (ST GEORGE)"
## [2] "LANDON CAMERON"
## [1] "multiple meet class req"
## [1] "NATHAN W DAVIS DPM (MURRAY)" "NATHAN W DAVIS"
## [1] "multiple meet class req"
## [1] "DR BRIAN RICHMAN (LAYTON)" "BRIAN H RICHMAN"
## [1] "multiple meet class req"
## [1] "CORAL DESERT FOOT ANKLE (ST GEORGE)"
## [2] "LANDON CAMERON"
## [1] "multiple meet class req"
## [1] "D ALDEN YATES DPM (ST GEORGE)" "DON A YATES"
## [1] "multiple meet class req"
## [1] "D ALDEN YATES DPM (ST GEORGE)" "DON A YATES"
## [1] "multiple meet class req"
## [1] "CORAL DESERT FOOT ANKLE (ST GEORGE)"
## [2] "LANDON CAMERON"
## [1] "multiple meet class req"
## [1] "SCRODCO P (SOUTHWEST FOOT & ANKLE)" "DAVID T MAGNESEN"
## [1] "multiple meet class req"
## [1] "CORAL DESERT FOOT ANKLE (ST GEORGE)"
## [2] "JEFFREY LEE STEWART"
## [1] "multiple meet class req"
## [1] "CORAL DESERT FOOT ANKLE (ST GEORGE)"
## [2] "JEFFREY LEE STEWART"
## [1] "multiple meet class req"
## [1] "DONALD R CALL JR DPM (SOUTH JORDAN)"
## [2] "DONALD R CALL"
## [1] "multiple meet class req"
## [1] "L CRAIG LARSEN DPM (MURRAY)" "CLARK C LARSEN"
## [1] "multiple meet class req"
## [1] "D ALDEN YATES DPM (ST GEORGE)" "DON A YATES"
## [1] "multiple meet class req"
## [1] "D ALDEN YATES DPM (ST GEORGE)" "DON A YATES"
## [1] "multiple meet class req"
## [1] "DR BRIAN RICHMAN (LAYTON)" "BRIAN H RICHMAN"
## [1] "multiple meet class req"
## [1] "L CRAIG LARSEN DPM (MURRAY)" "CLARK C LARSEN"
## [1] "multiple meet class req"
## [1] "DR BRIAN RICHMAN (LAYTON)" "BRIAN H RICHMAN"
## [1] "multiple meet class req"
## [1] "CANYON FOOT AND ANKLE (SPANISH FORK)"
## [2] "LEVI J BERRY"
## [1] "multiple meet class req"
## [1] "ANDREW H BURGON PODIATRY (SUMMIT FOOT AND ANKLE)"
## [2] "ANDREW HARRIS BURGON"
## [1] "multiple meet class req"
## [1] "D ALDEN YATES DPM (ST GEORGE)" "DON A YATES"
## [1] "multiple meet class req"
## [1] "CORAL DESERT FOOT ANKLE (ST GEORGE)"
## [2] "LANDON CAMERON"
## [1] "multiple meet class req"
## [1] "DR BRIAN RICHMAN (LAYTON)" "BRIAN H RICHMAN"
## [1] "multiple meet class req"
## [1] "D ALDEN YATES DPM (ST GEORGE)" "DON A YATES"
## [1] "multiple meet class req"
## [1] "DR BRIAN RICHMAN (LAYTON)" "BRIAN H RICHMAN"
## [1] "multiple meet class req"
## [1] "K2 FOOT AND ANKLE P (MURRAY)" "STANTON MILLER SMITH"
## [1] "multiple meet class req"
## [1] "DAVID H WARBY DPM (LAYTON)" "DAVID H WARBY"
## [1] "multiple meet class req"
## [1] "CORAL DESERT FOOT ANKLE (ST GEORGE)"
## [2] "JEFFREY LEE STEWART"
## [1] "multiple meet class req"
## [1] "D ALDEN YATES DPM (ST GEORGE)" "DON A YATES"
## [1] "multiple meet class req"
## [1] "CORAL DESERT FOOT ANKLE (ST GEORGE)"
## [2] "LANDON CAMERON"
## [1] "multiple meet class req"
## [1] "CORAL DESERT FOOT ANKLE (ST GEORGE)"
## [2] "LANDON CAMERON"
## [1] "multiple meet class req"
## [1] "K2 FOOT AND ANKLE P (MURRAY)" "STANTON MILLER SMITH"toe_nail_n_visit_clean_btbv5$tp_med_med %>% summary()## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.0 204.7 221.9 254.8 279.6 1599.9toe_q1 <- toe_nail_n_visit_clean_btbv5$tp_med_med %>% quantile(.25)
toe_q3 <- toe_nail_n_visit_clean_btbv5$tp_med_med %>% quantile(.75)
toe_iqr <- toe_nail_n_visit_clean_btbv5$tp_med_med %>% IQR()toe_nail_n_visit_clean_btbv5_final <- toe_nail_n_visit_clean_btbv5[tp_med_med >toe_q1 -1.5*toe_iqr & tp_med_med < toe_q3 + 1.5*toe_iqr ]toe_nail_n_visit_clean_btbv5$tp_med_med %>% summary()## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.0 204.7 221.9 254.8 279.6 1599.9toe_nail_n_visit_clean_btbv5_final$tp_med_med %>% summary()## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 106.0 204.7 221.3 237.2 272.8 383.0bq_table_upload(x=procedure_dev_table, values= toe_nail_n_visit_clean_btbv5_final, create_disposition='CREATE_IF_NEEDED', write_disposition='WRITE_APPEND')