Hernia Repair
Introduction
In this section we will: * identify which Hernia Repair procedures were excluded and why * identify which different Hernia Repair subgroups were created and why
If you have questions or concerns about this data please contact Alexander Nielson (alexnielson@utah.gov)
Load Libraries
Load Libraries
library(data.table)
library(tidyverse)## Warning: replacing previous import 'vctrs::data_frame' by 'tibble::data_frame'
## when loading 'dplyr'library(stringi)
library(ggridges)
library(broom)
library(disk.frame)
library(RecordLinkage)
library(googlesheets4)
library(bigrquery)
library(DBI)
devtools::install_github("utah-osa/hcctools2", upgrade="never" )
library(hcctools2)Establish color palettes
cust_color_pal1 <- c(
"Anesthesia" = "#f3476f",
"Facility" = "#e86a33",
"Medicine Proc" = "#e0a426",
"Pathology" = "#77bf45",
"Radiology" = "#617ff7",
"Surgery" = "#a974ee"
)
cust_color_pal2 <- c(
"TRUE" = "#617ff7",
"FALSE" = "#e0a426"
)
cust_color_pal3 <- c(
"above avg" = "#f3476f",
"avg" = "#77bf45",
"below avg" = "#a974ee"
)
fac_ref_regex <- "(UTAH)|(IHC)|(HOSP)|(HOSPITAL)|(CLINIC)|(ANESTH)|(SCOPY)|(SURG)|(LLC)|(ASSOC)|(MEDIC)|(CENTER)|(ASSOCIATES)|(U OF U)|(HEALTH)|(OLOGY)|(OSCOPY)|(FAMILY)|(VAMC)|(SLC)|(SALT LAKE)|(CITY)|(PROVO)|(OGDEN)|(ENDO )|( VALLEY)|( REGIONAL)|( CTR)|(GRANITE)|( INSTITUTE)|(INSTACARE)|(WASATCH)|(COUNTY)|(PEDIATRIC)|(CORP)|(CENTRAL)|(URGENT)|(CARE)|(UNIV)|(ODYSSEY)|(MOUNTAINSTAR)|( ORTHOPEDIC)|(INSTITUT)|(PARTNERSHIP)|(PHYSICIAN)|(CASTLEVIEW)|(CONSULTING)|(MAGEMENT)|(PRACTICE)|(EMERGENCY)|(SPECIALISTS)|(DIVISION)|(GUT WHISPERER)|(INTERMOUNTAIN)|(OBGYN)"Connect to GCP database
bigrquery::bq_auth(path = 'D:/gcp_keys/healthcare-compare-prod-95b3b7349c32.json')
# set my project ID and dataset name
project_id <- 'healthcare-compare-prod'
dataset_name <- 'healthcare_compare'
con <- dbConnect(
bigrquery::bigquery(),
project = project_id,
dataset = dataset_name,
billing = project_id
)Get NPI table
query <- paste0("SELECT npi, clean_name, osa_group, osa_class, osa_specialization
FROM `healthcare-compare-prod.healthcare_compare.npi_full`")
#bq_project_query(billing, query) # uncomment to determine billing price for above query.
npi_full <- dbGetQuery(con, query) %>%
data.table()get a subset of the NPI providers based upon taxonomy groups
gs4_auth(email="alexnielson@utah.gov")surgery <- read_sheet("https://docs.google.com/spreadsheets/d/1GY8lKwUJuPHtpUl4EOw9eriLUDG9KkNWrMbaSnA5hOU/edit#gid=0",
sheet="major_surgery") %>% as.data.table## Auto-refreshing stale OAuth token.## Reading from "Doctor Types to Keep"## Range "'major_surgery'"surgery <- surgery[is.na(Remove) ] %>% .[["NUCC Classification"]] npi_prov_pair <- npi_full[osa_class %in% surgery] %>%
.[,.(npi=npi,
clean_name = clean_name
)
]Load Data
bun_proc <- disk.frame("full_apcd.df")hernia <- bun_proc[surg_bun_descr %>% stri_detect_regex("hern")]hernia <- hernia[,`:=`(
surg_sp_name_clean = surg_sp_npi %>% map_chr(get_npi_standard_name),
surg_bp_name_clean = surg_bp_npi %>% map_chr(get_npi_standard_name),
medi_sp_name_clean = medi_sp_npi %>% map_chr(get_npi_standard_name),
medi_bp_name_clean = medi_bp_npi %>% map_chr(get_npi_standard_name),
radi_sp_name_clean = radi_sp_npi %>% map_chr(get_npi_standard_name),
radi_bp_name_clean = radi_bp_npi %>% map_chr(get_npi_standard_name),
path_sp_name_clean = path_sp_npi %>% map_chr(get_npi_standard_name),
path_bp_name_clean = path_bp_npi %>% map_chr(get_npi_standard_name),
anes_sp_name_clean = anes_sp_npi %>% map_chr(get_npi_standard_name),
anes_bp_name_clean = anes_bp_npi %>% map_chr(get_npi_standard_name),
faci_sp_name_clean = faci_sp_npi %>% map_chr(get_npi_standard_name),
faci_bp_name_clean = faci_bp_npi %>% map_chr(get_npi_standard_name)
)]hernia %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.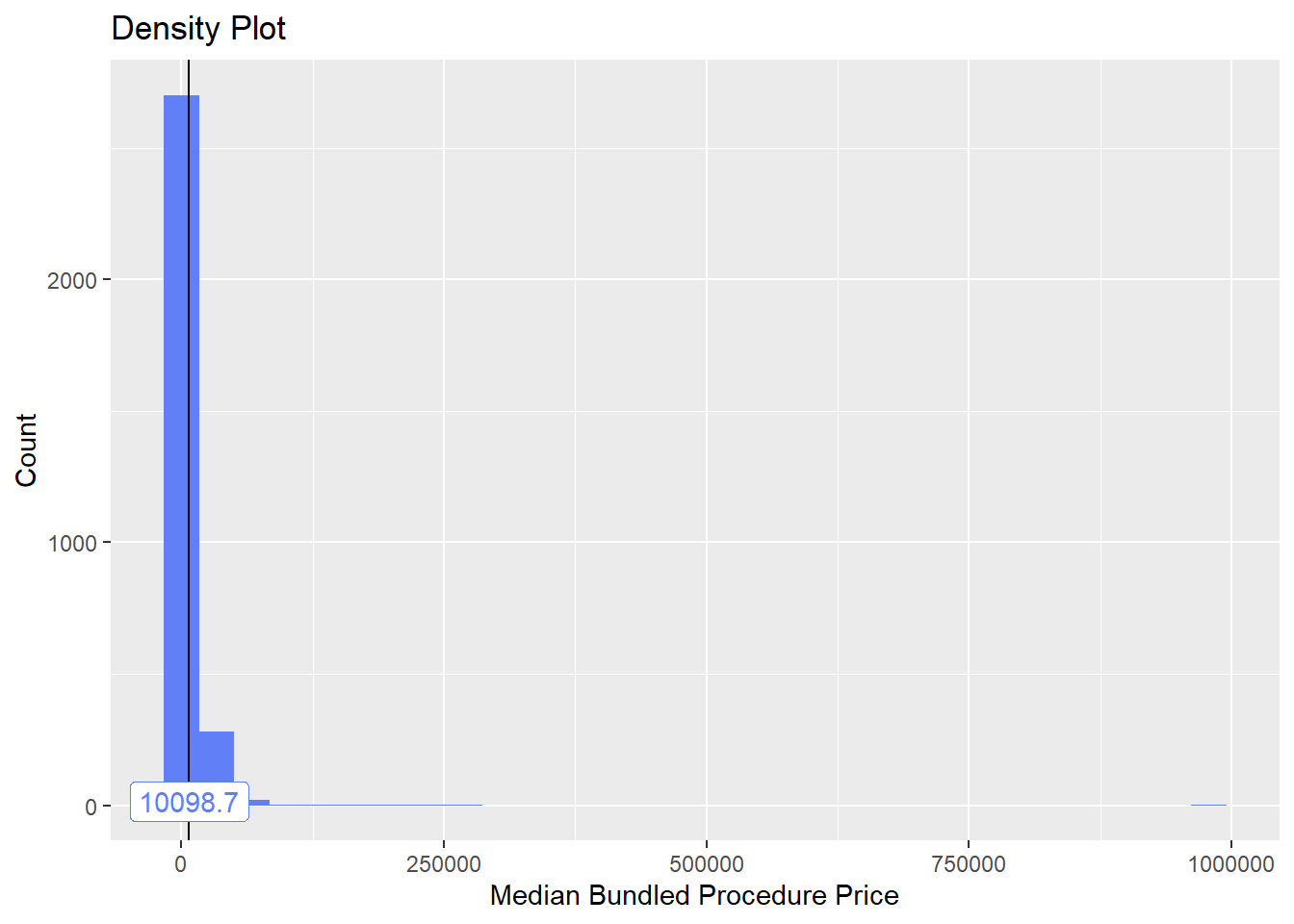
hernia %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 465 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.876
## 2 duration_mean 0.685
## 3 surg_bun_t_imag 0.652
## 4 surg_bun_t_fluid 0.612
## 5 medi_bun_t_subseq 0.596
## 6 radi_bun_t_head 0.55
## 7 medi_bun_t_with 0.504
## 8 medi_bun_t_eye_exam 0.460
## 9 radi_bun_t_upper 0.441
## 10 surg_bun_t_cath 0.419
## # ... with 455 more rowshernia %>% plot_price_vs_duration() %>% print()
hernia <- hernia[duration_mean<2 & tp_med > 0 & tp_med < 50000 & faci_bun_sum_med > 500]hernia %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
hernia %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 411 x 2
## name correlation
## <chr> <dbl>
## 1 duration_mean 0.5
## 2 surg_bun_t_lap 0.382
## 3 anes_bun_t_upper 0.352
## 4 anes_bun_t_hernia 0.342
## 5 anes_bun_t_repair 0.339
## 6 surg_bun_t_repair 0.338
## 7 surg_bun_t_esoph 0.336
## 8 anes_bun_t_surg 0.333
## 9 medi_bun_t_inject 0.329
## 10 anes_bun_t_abdom 0.305
## # ... with 401 more rowshernia %>% get_tag_density_information("surg_bun_t_lap") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_lap"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 795## $dist_plots
##
## $stat_tables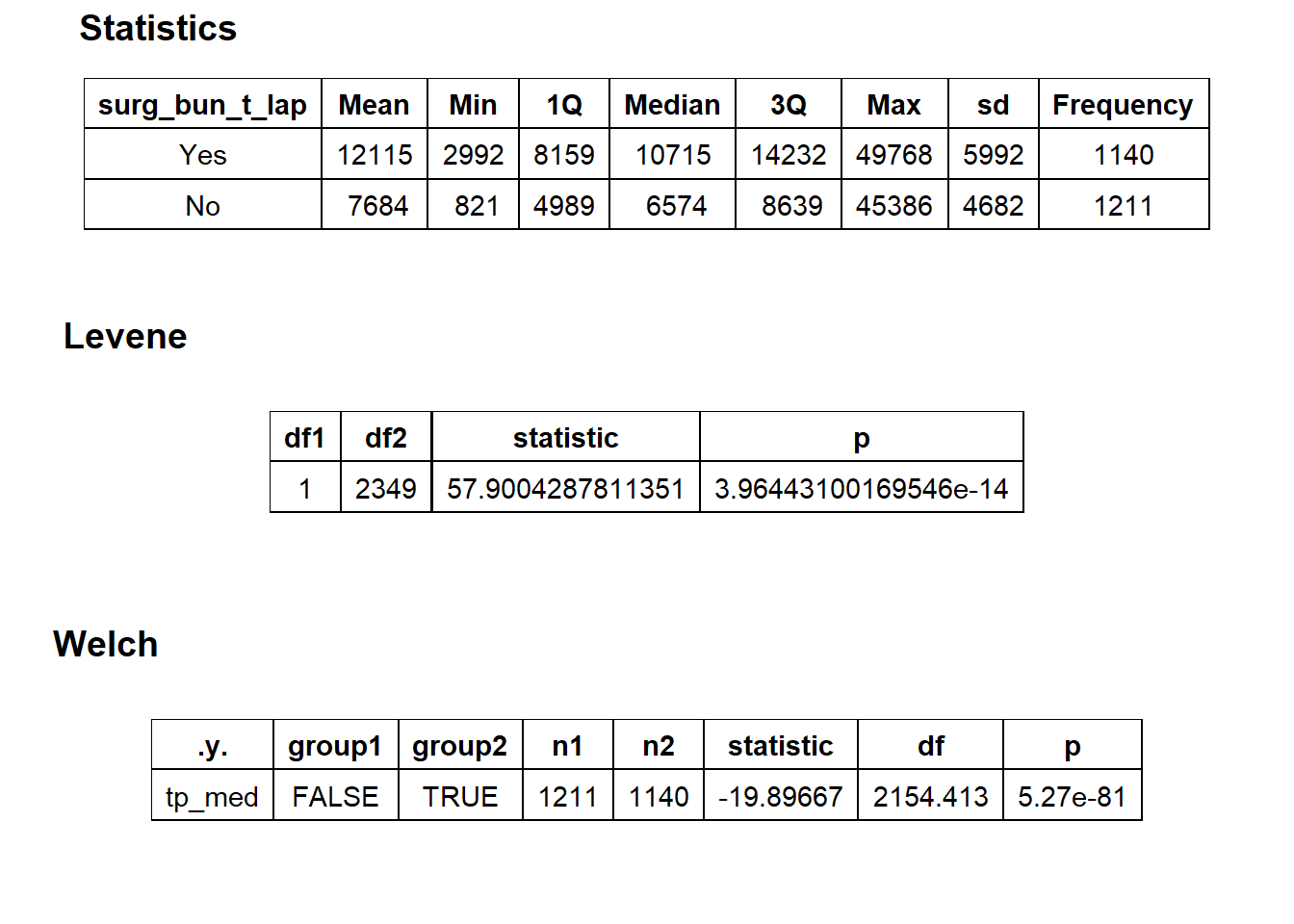
hernia_lap <- hernia[surg_bun_t_lap ==T]
hernia_n_lap <- hernia[surg_bun_t_lap==F]hernia_lap
hernia_lap %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.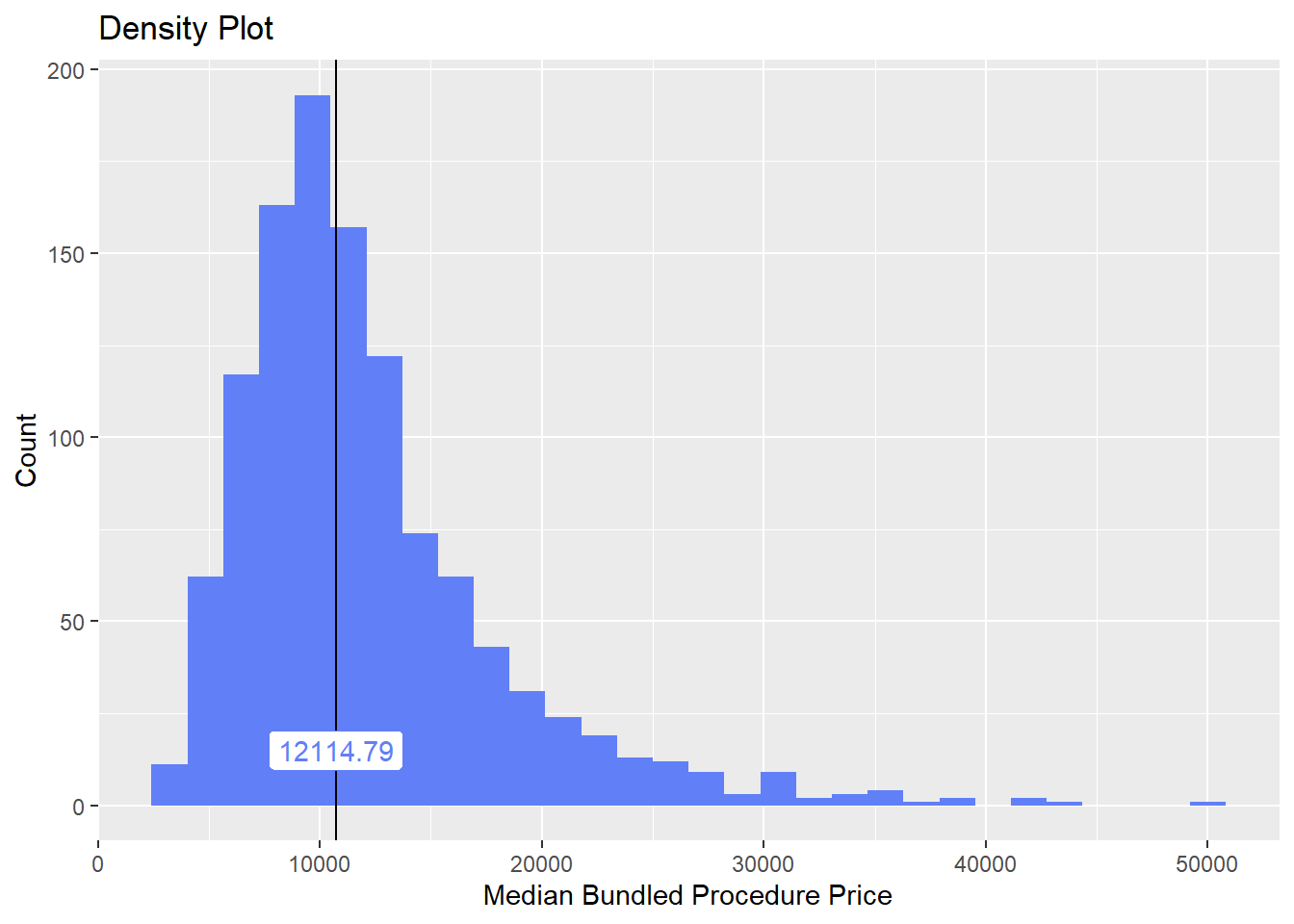
hernia_lap %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 306 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.516
## 2 duration_mean 0.505
## 3 medi_bun_t_sodium 0.349
## 4 medi_bun_t_inject 0.348
## 5 surg_bun_t_esoph 0.336
## 6 anes_bun_t_lower 0.335
## 7 surg_bun_t_hernia 0.303
## 8 surg_bun_t_art 0.282
## 9 surg_bun_t_partial 0.278
## 10 anes_bun_t_upper 0.276
## # ... with 296 more rowshernia_lap %>% get_tag_density_information("medi_bun_t_sodium" ) %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_sodium"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 1850## $dist_plots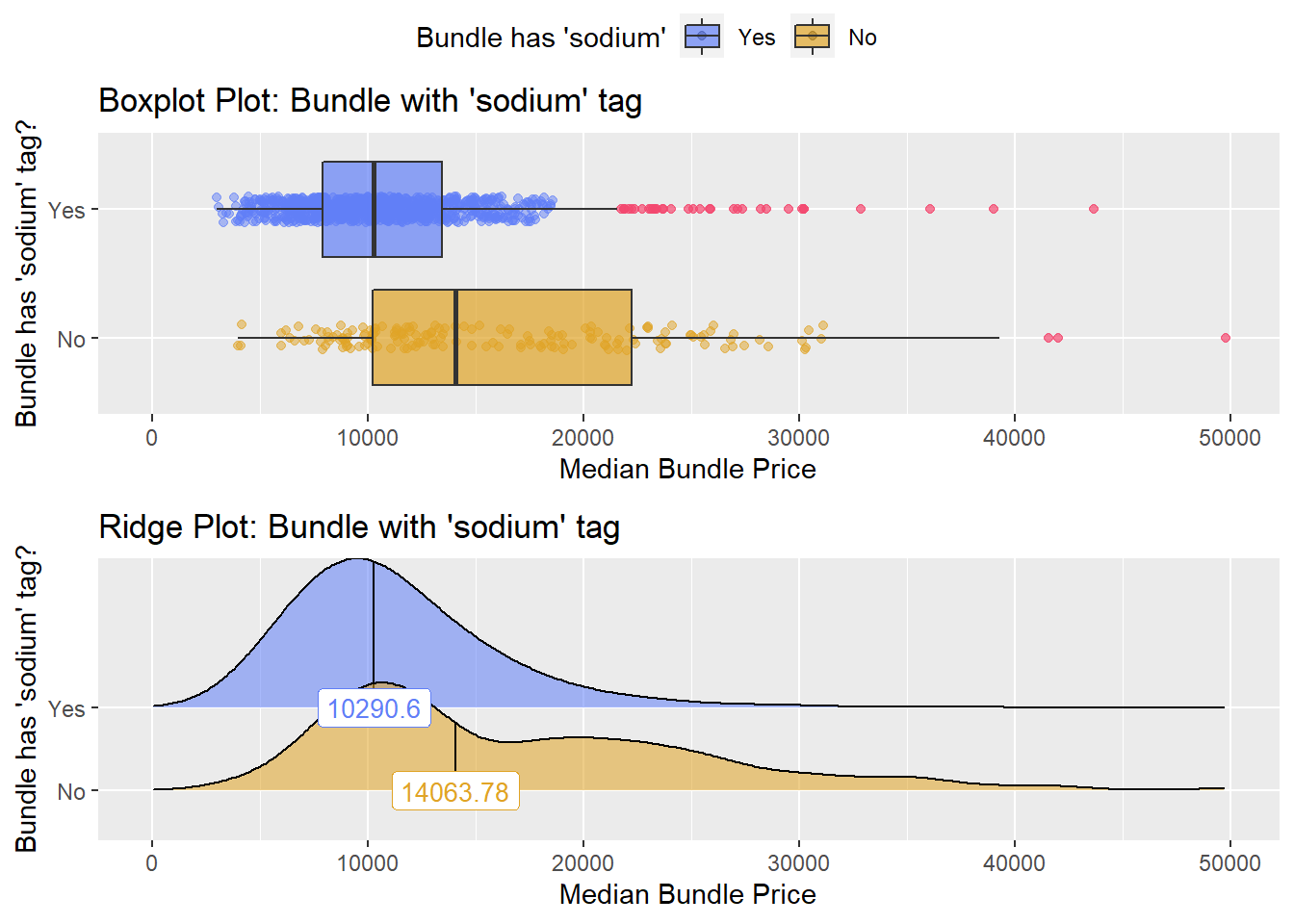
##
## $stat_tables
hernia_lap <- hernia_lap[medi_bun_t_sodium == T]hernia_lap %>% get_tag_density_information("surg_bun_t_esoph" ) %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_esoph"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 1190## $dist_plots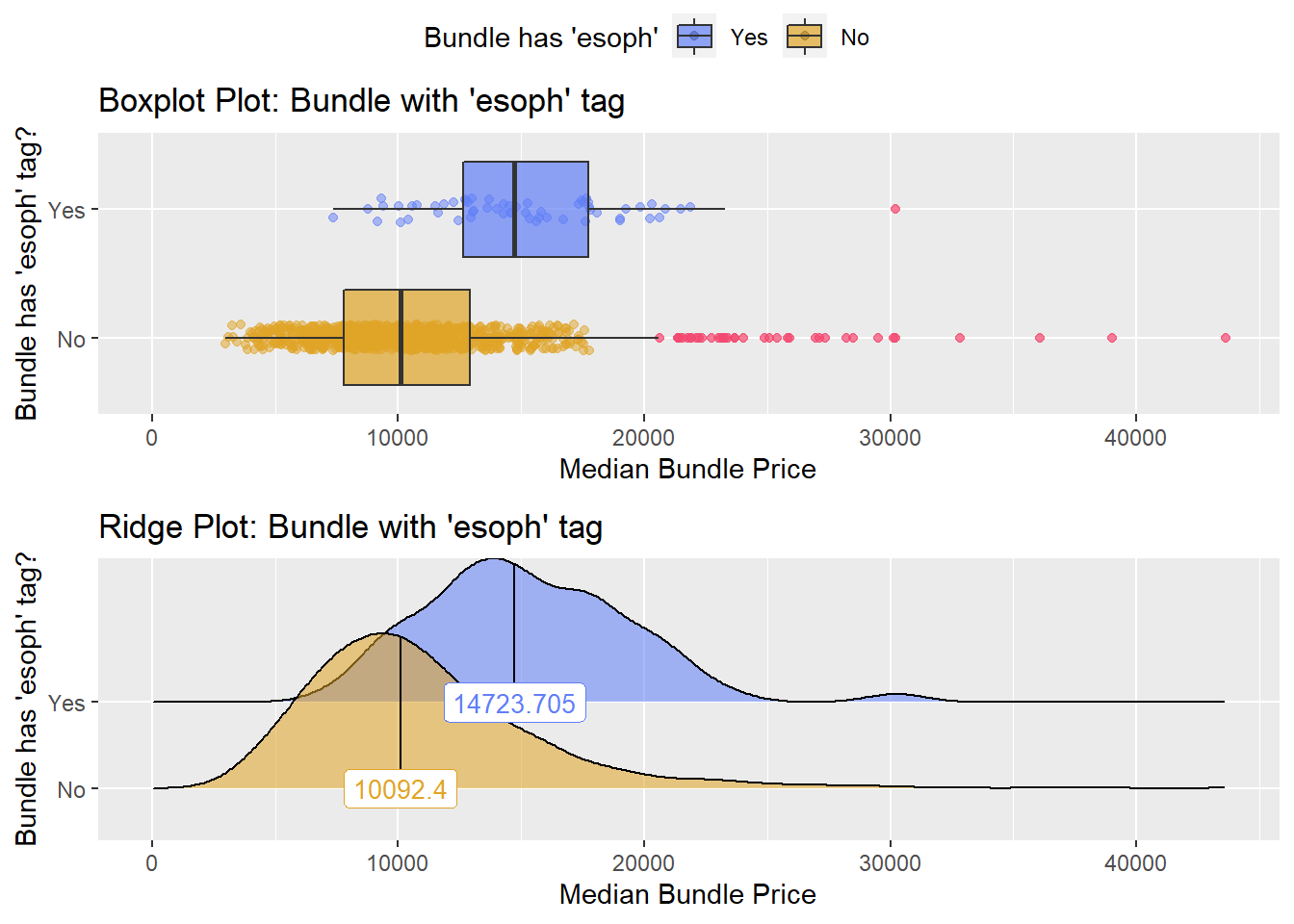
##
## $stat_tables
hernia_lap <- hernia_lap[surg_bun_t_esoph==F]hernia_lap %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.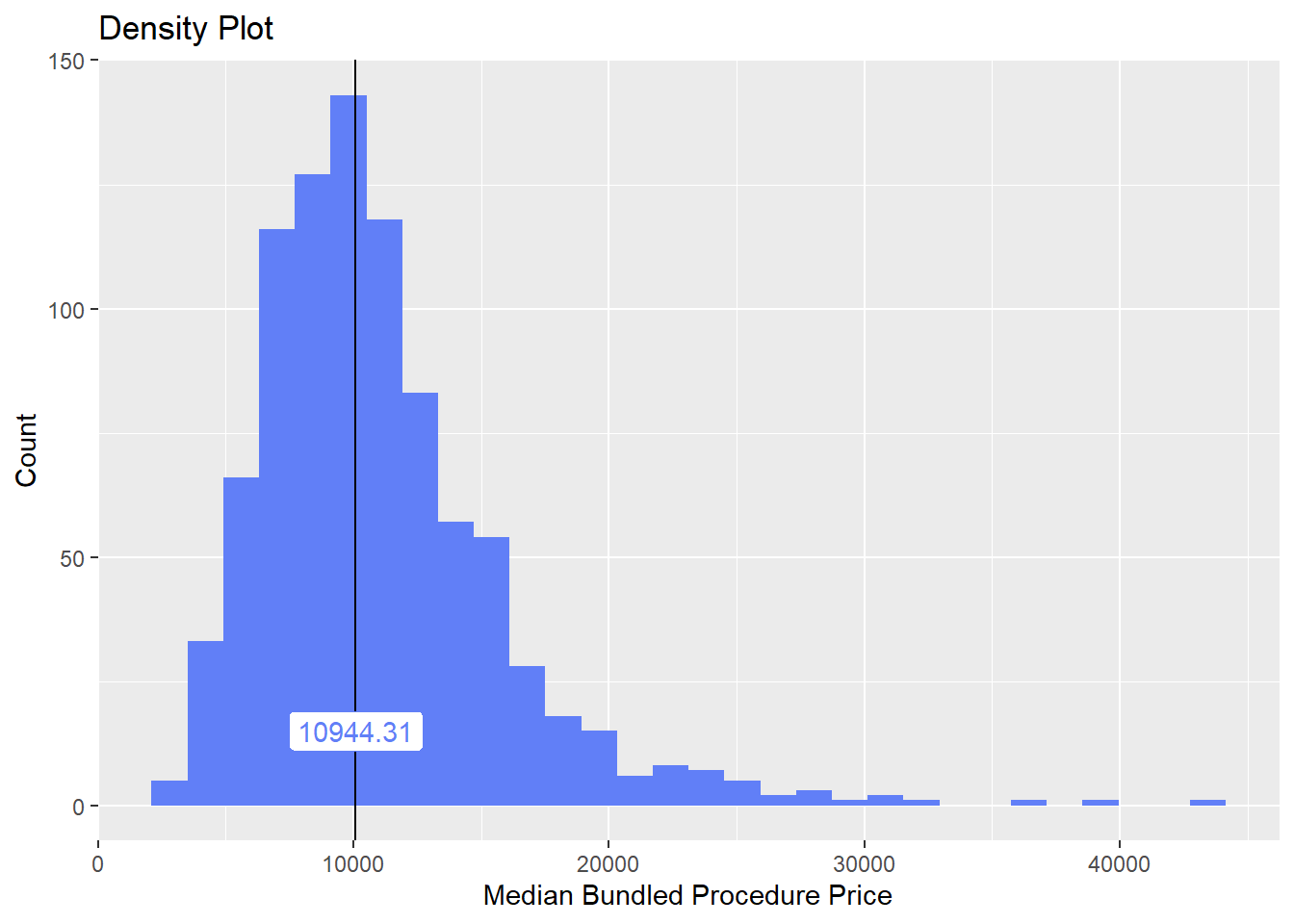
hernia_lap %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 224 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_lymph 0.286
## 2 surg_bun_t_urethra 0.286
## 3 surg_bun_t_prostat 0.280
## 4 surg_bun_t_prostate 0.280
## 5 surg_bun_t_bladder 0.27
## 6 surg_bun_t_injection 0.246
## 7 surg_bun_t_block 0.237
## 8 anes_bun_t_lower 0.230
## 9 surg_bun_t_scope 0.228
## 10 surg_bun_t_stone 0.228
## # ... with 214 more rowshernia_lap %>% get_tag_density_information("surg_bun_t_lymph") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_lymph"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 3950## $dist_plots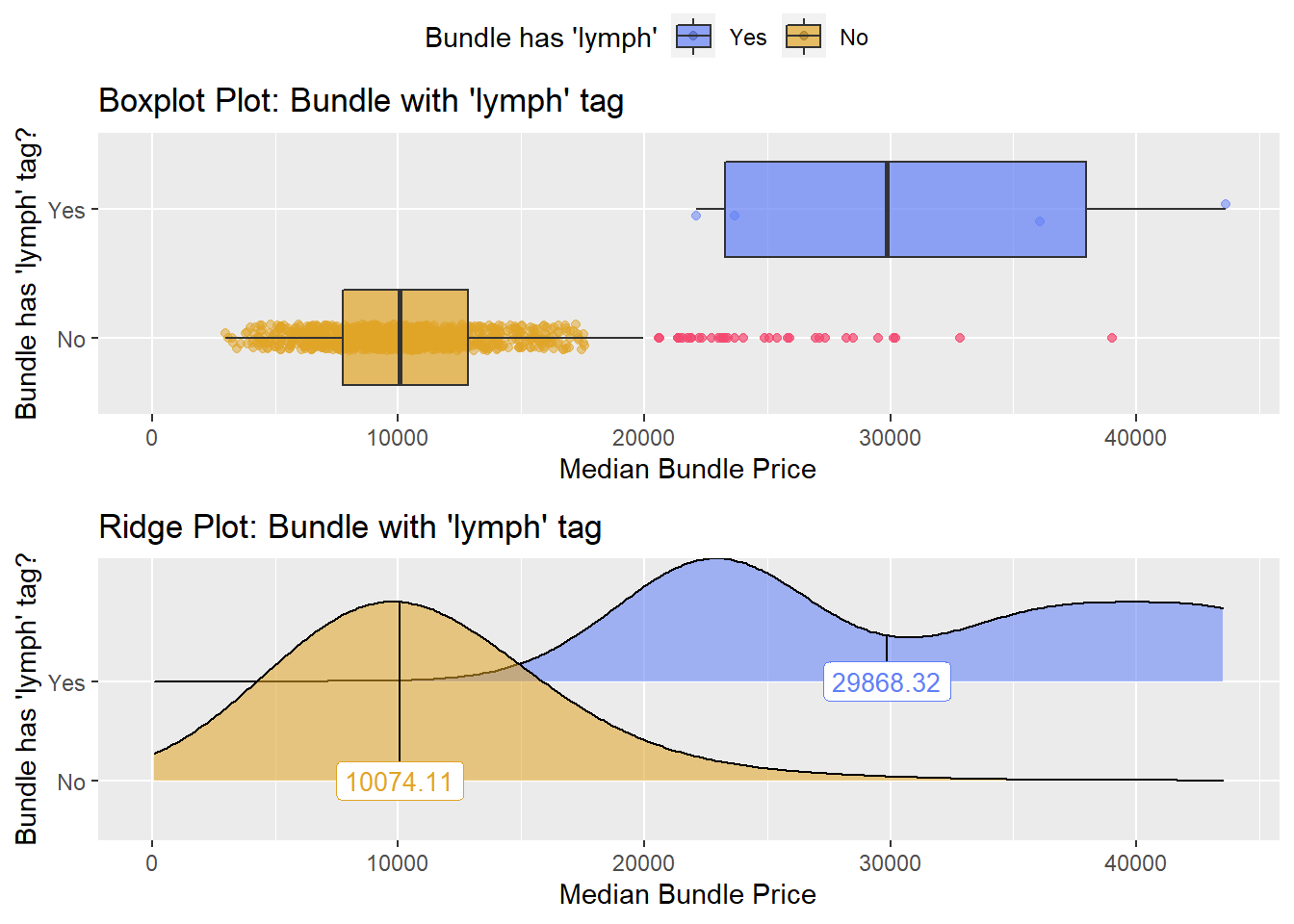
##
## $stat_tables
hernia_lap <- hernia_lap[surg_bun_t_lymph==F]hernia_lap %>% get_tag_density_information("surg_bun_t_urethra") %>% print()hernia_lap <- hernia_lap[surg_bun_t_urethra==F]hernia_lap %>% get_tag_density_information("surg_bun_t_prostate") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_prostate"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 1670## $dist_plots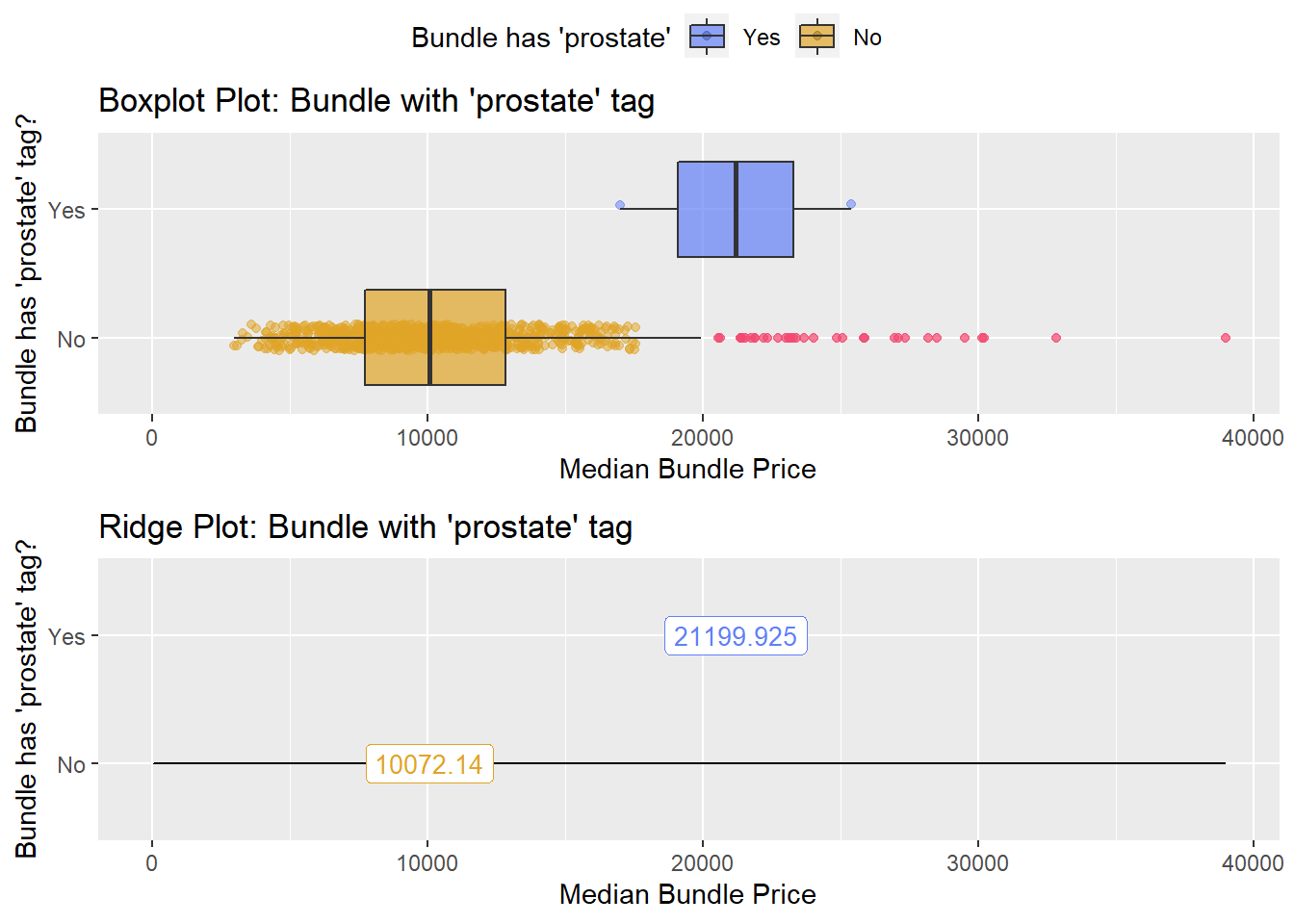
##
## $stat_tables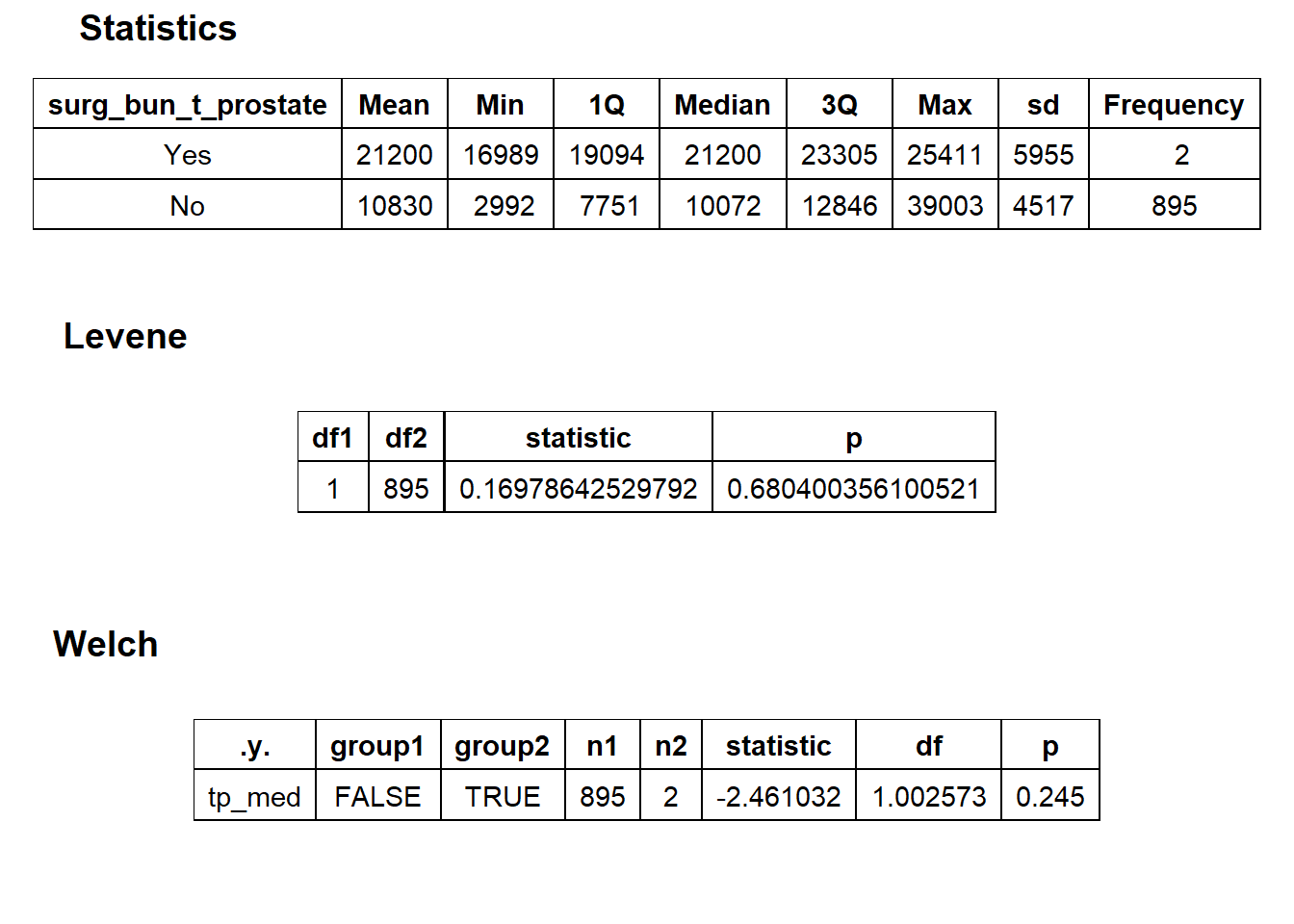
hernia_lap <- hernia_lap[surg_bun_t_prostate==F]hernia_lap %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
hernia_lap %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 215 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_injection 0.256
## 2 surg_bun_t_graft 0.243
## 3 surg_bun_t_musc 0.243
## 4 surg_bun_t_block 0.234
## 5 surg_bun_t_skin 0.226
## 6 anes_bun_t_lower 0.208
## 7 anes_bun_t_upper 0.207
## 8 anes_bun_t_kidney 0.184
## 9 anes_bun_t_ureter 0.184
## 10 surg_bun_t_ant 0.167
## # ... with 205 more rowshernia_lap %>% get_tag_density_information("surg_bun_t_graft") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_graft"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 1770## $dist_plots
##
## $stat_tables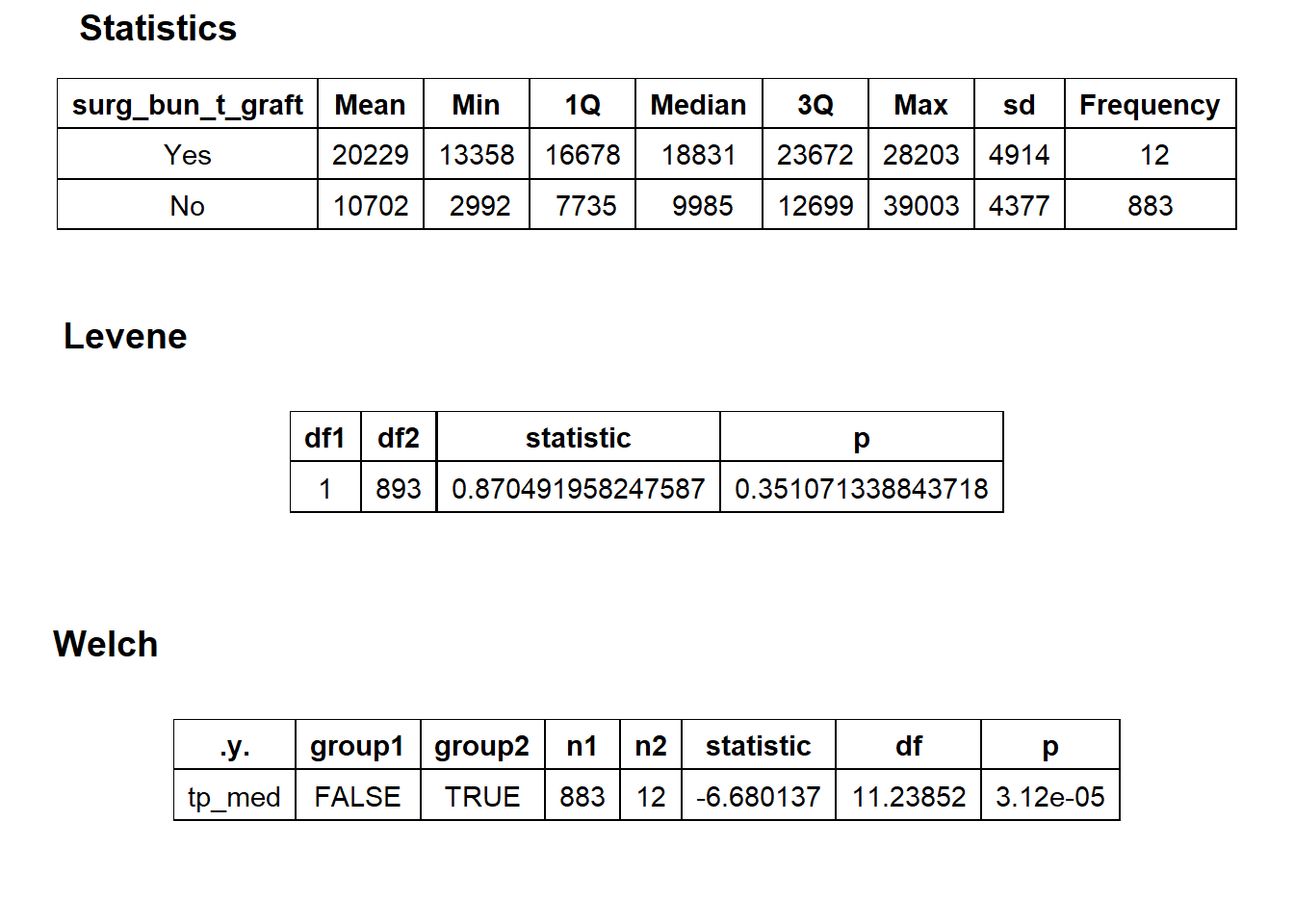
hernia_lap <- hernia_lap[surg_bun_t_graft==F]hernia_lap %>% get_tag_density_information("surg_bun_t_block") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_block"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 1180## $dist_plots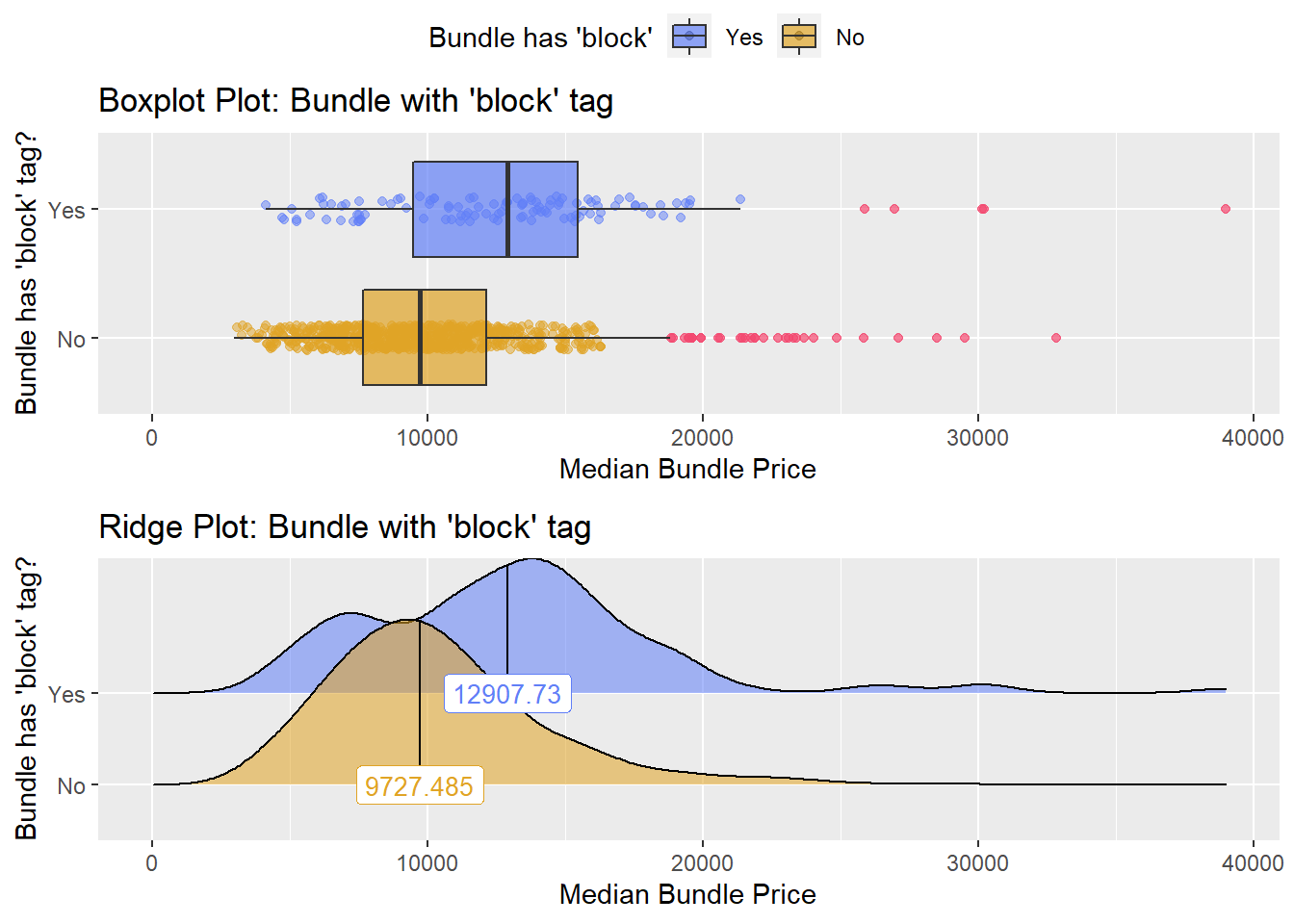
##
## $stat_tables
hernia_lap <- hernia_lap[surg_bun_t_block==F]# hernia_lap %>% get_tag_density_information("surg_bun_t_skin") %>% print()hernia_lap <- hernia_lap[surg_bun_t_skin==F]hernia_lap %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.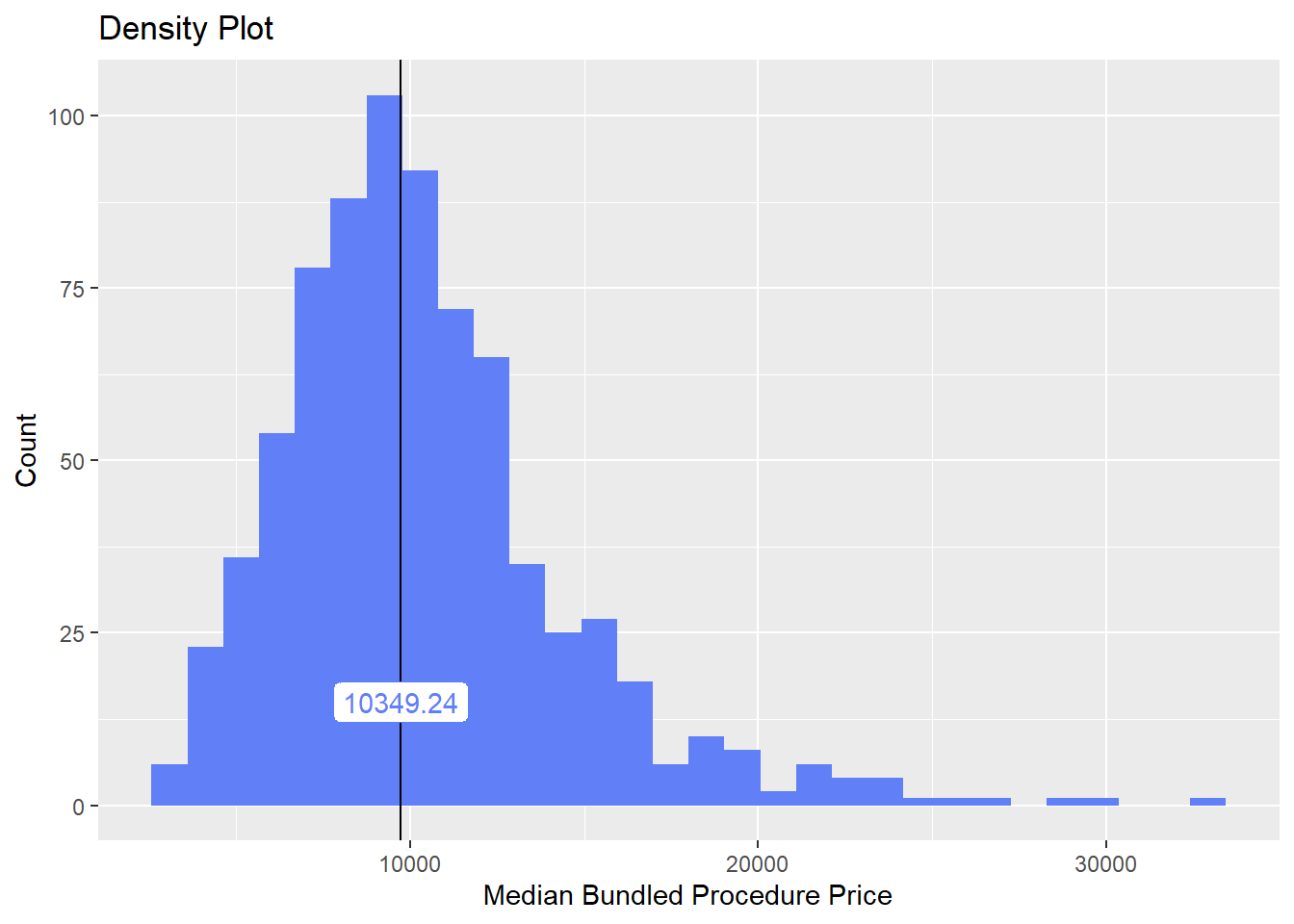
hernia_lap %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 186 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_kidney 0.227
## 2 anes_bun_t_ureter 0.227
## 3 surg_bun_t_ant 0.208
## 4 path_bun_t_total 0.203
## 5 surg_bun_t_drug 0.200
## 6 surg_bun_t_nephrectomy 0.200
## 7 surg_bun_t_implant 0.194
## 8 anes_bun_t_lower 0.192
## 9 path_bun_t_time 0.191
## 10 anes_bun_t_upper 0.187
## # ... with 176 more rows# hernia_lap %>% get_tag_density_information("surg_bun_t_injection") %>% print()
hernia_lap %>% get_tag_density_information("anes_bun_t_upper") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ anes_bun_t_upper"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 1310## $dist_plots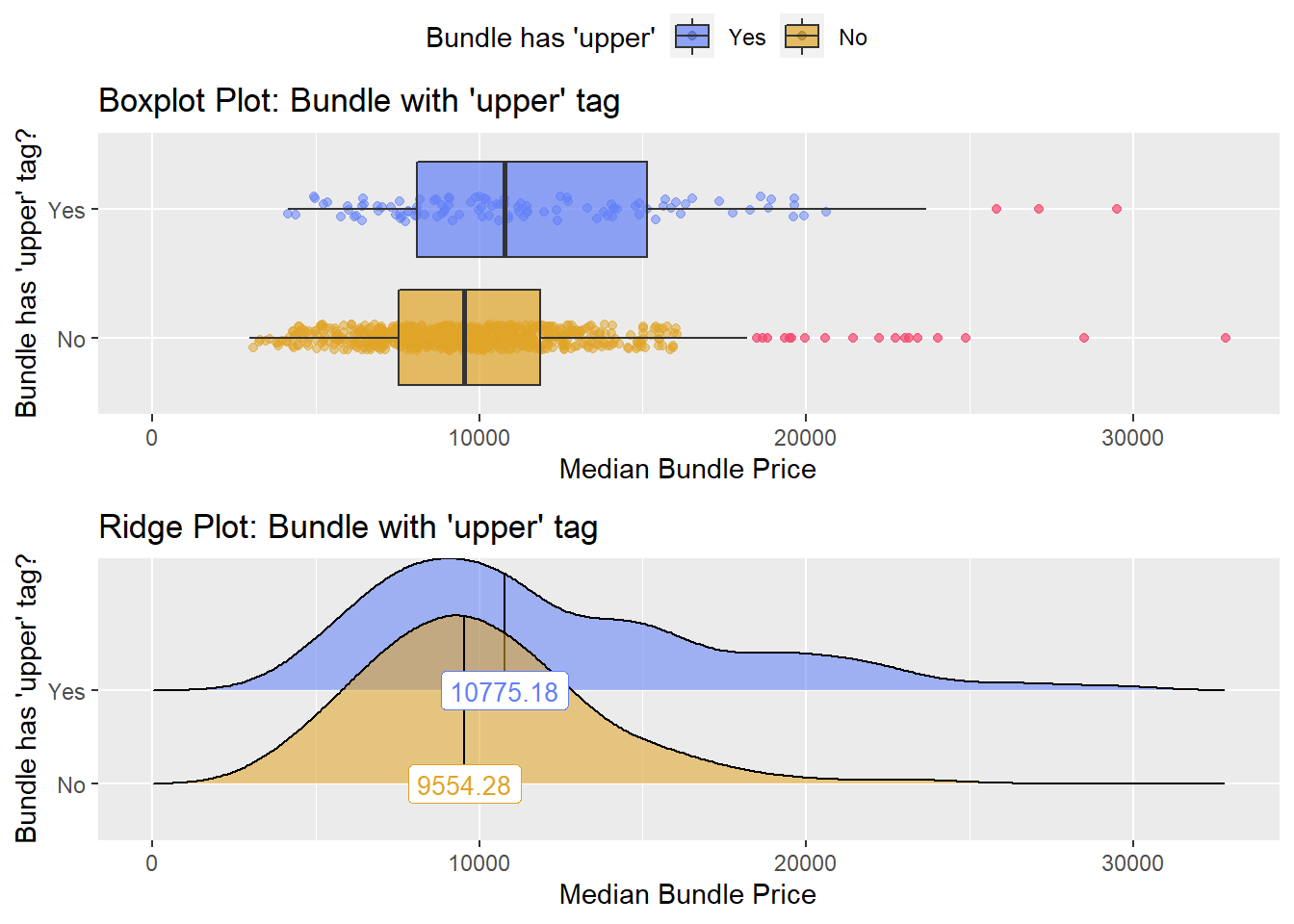
##
## $stat_tables
hernia_lap %>% get_tag_density_information("anes_bun_t_kidney") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.
## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ anes_bun_t_kidney"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 1680## $dist_plots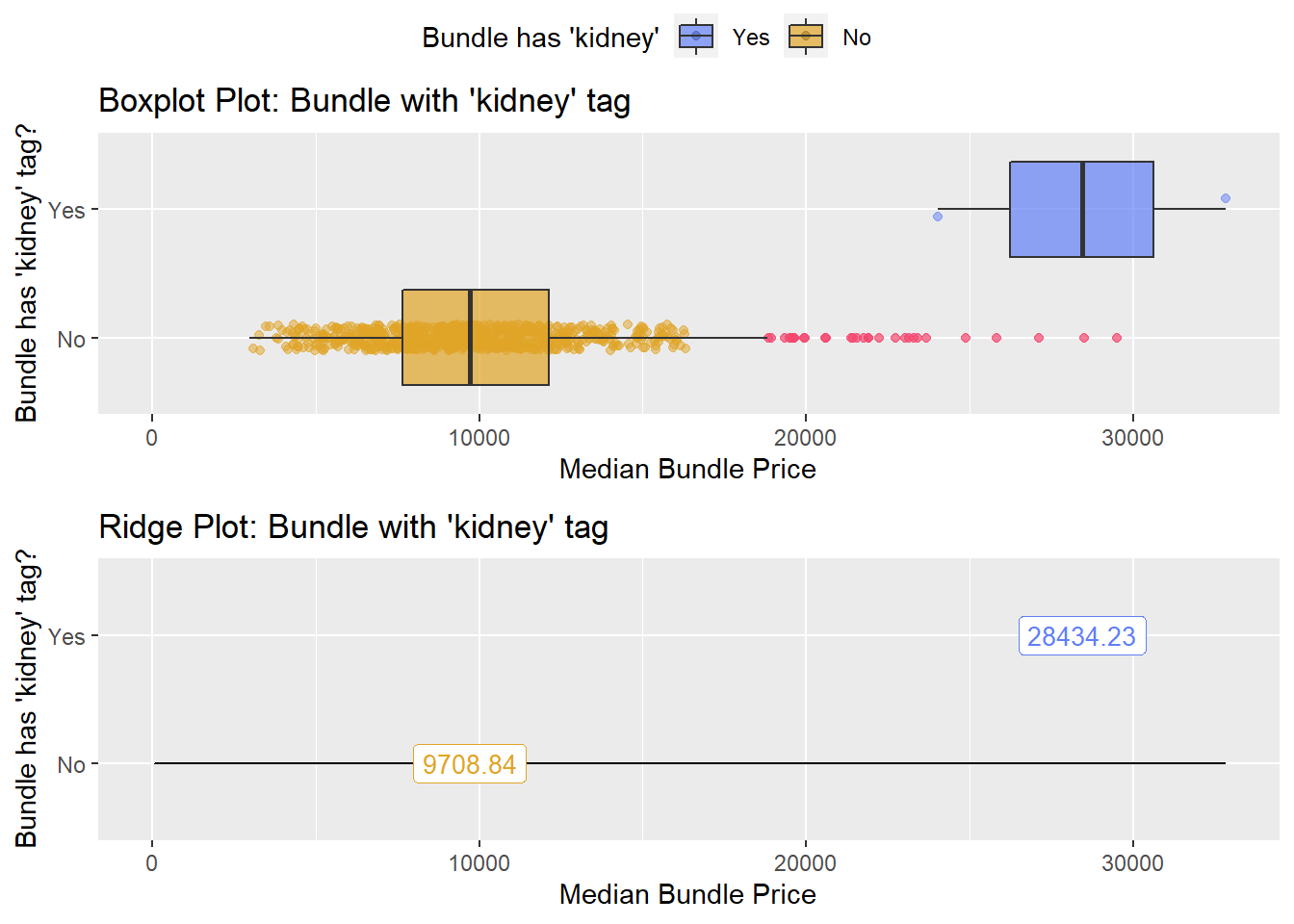
##
## $stat_tables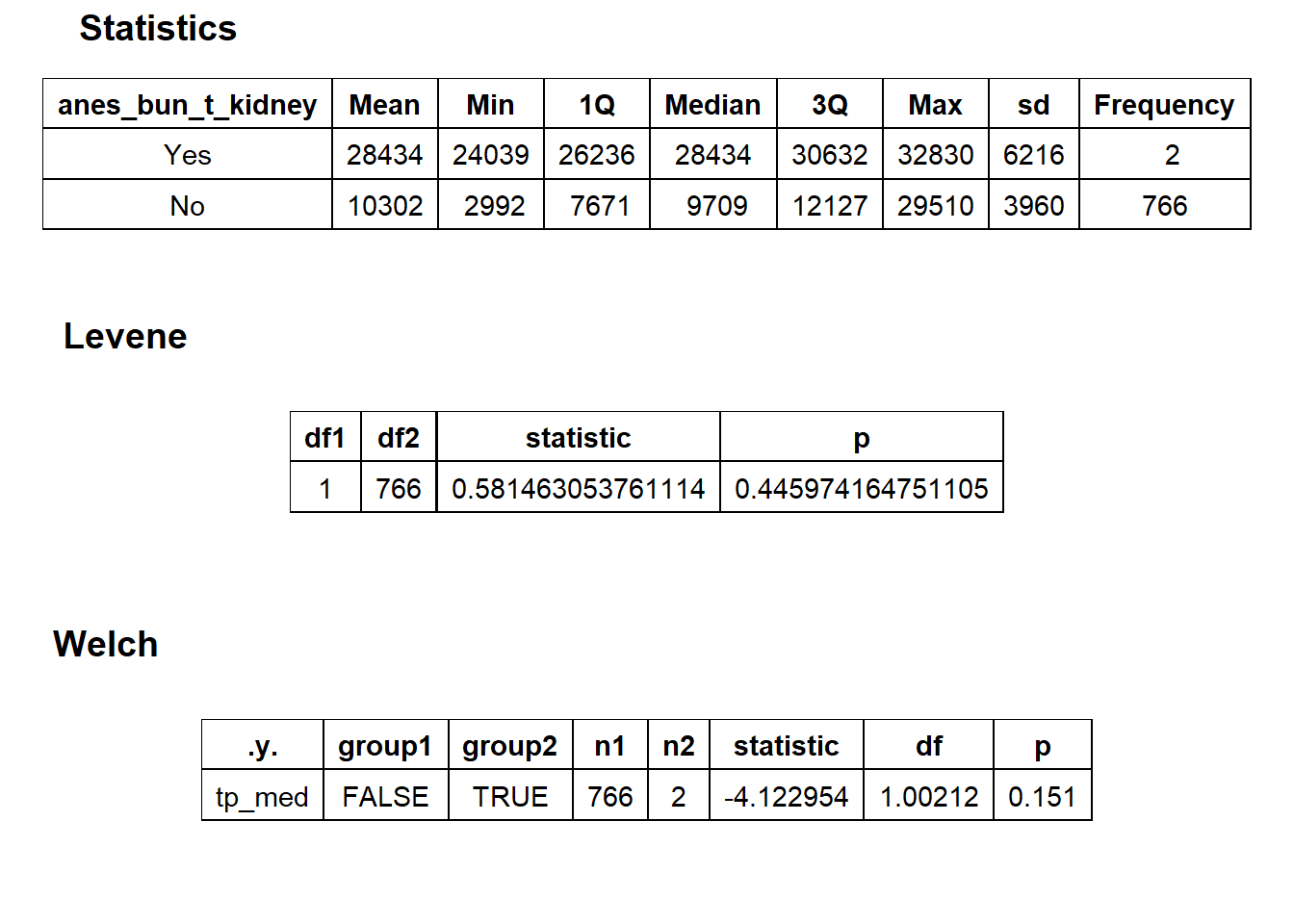
hernia_lap %>% get_tag_density_information("anes_bun_t_ureter") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.
## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ anes_bun_t_ureter"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 1680## $dist_plots
##
## $stat_tables
hernia_lap %>% get_tag_density_information("surg_bun_t_ant") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.
## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_ant"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 2300## $dist_plots
##
## $stat_tables
hernia_lap %>% get_tag_density_information("path_bun_t_complete") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.
## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_complete"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 1170## $dist_plots
##
## $stat_tables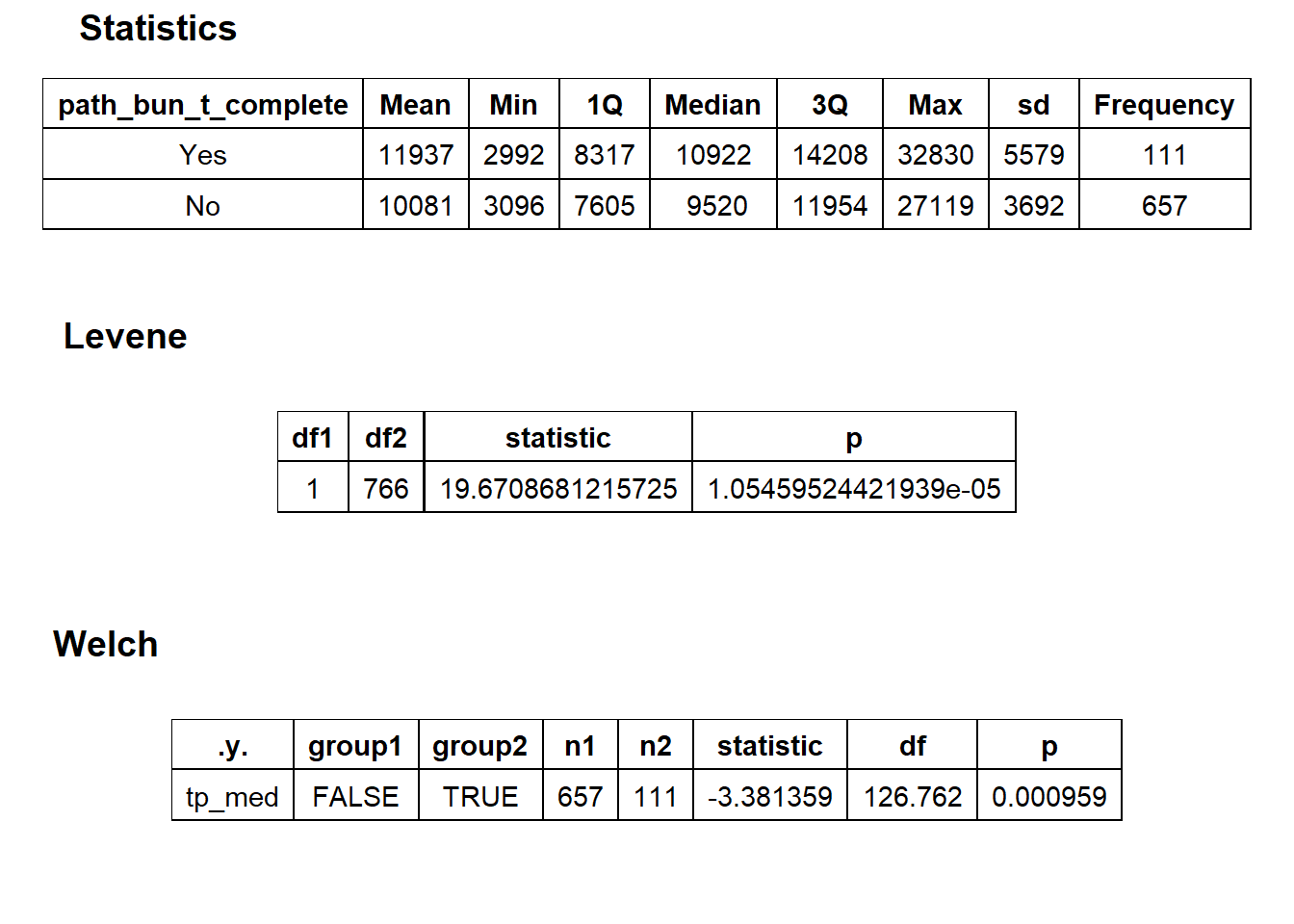
hernia_lap <- hernia_lap[surg_bun_t_injection ==F & anes_bun_t_kidney == F & anes_bun_t_ureter==F & surg_bun_t_ant==F ]hernia_lap %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.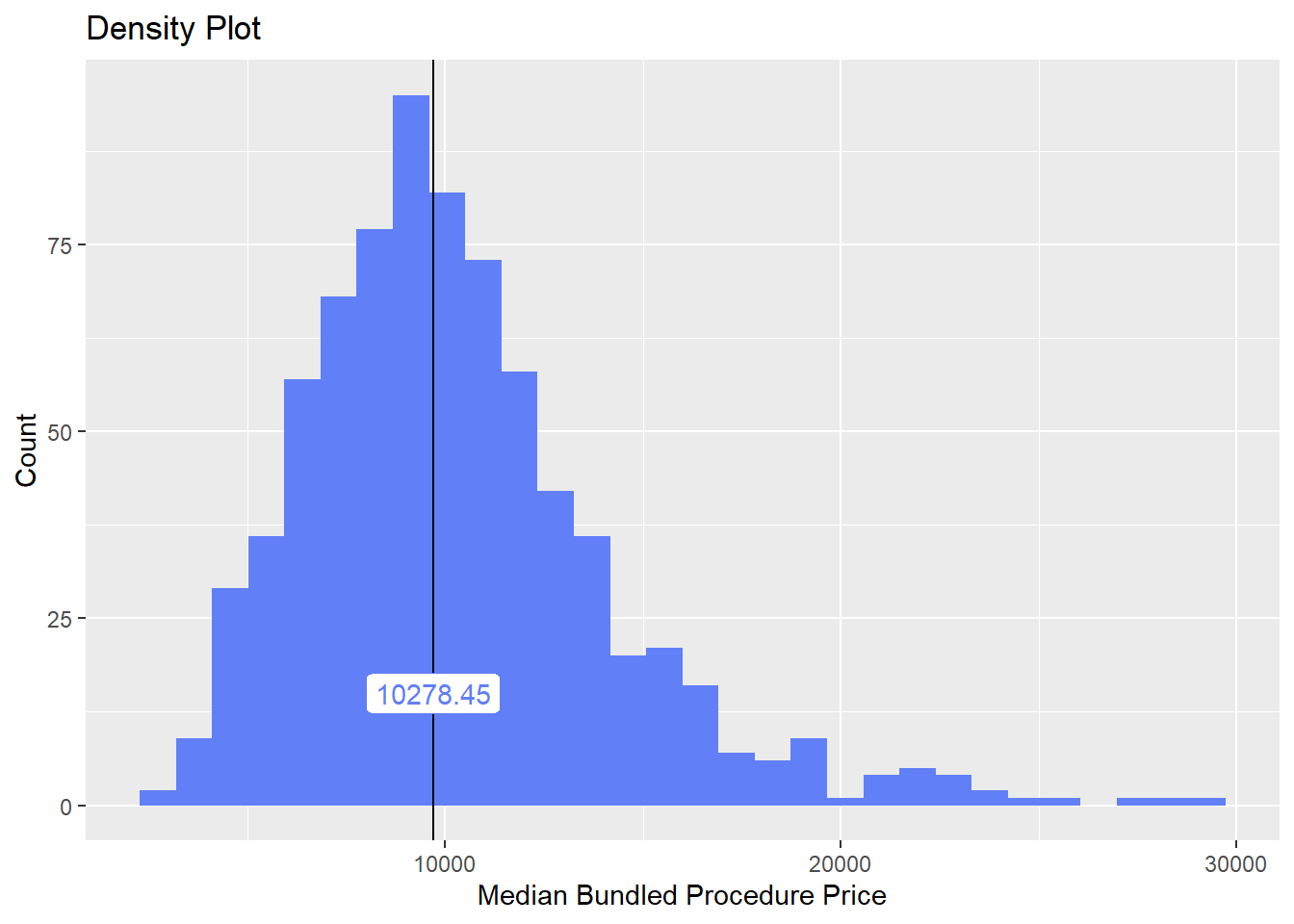
hernia_lap %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 173 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_upper 0.193
## 2 anes_bun_t_lower 0.176
## 3 path_bun_t_total 0.169
## 4 surg_bun_t_abd 0.150
## 5 path_bun_t_complete 0.140
## 6 path_bun_t_magnesium 0.123
## 7 path_bun_t_phosphorus 0.123
## 8 surg_bun_t_uterus 0.121
## 9 path_bun_t_time 0.109
## 10 path_bun_t_metabolic 0.107
## # ... with 163 more rowshernia_lap = surg_bun_t_uters
hernia_lap <- hernia_lap[surg_bun_t_uterus == F & surg_bun_t_uterus==F ]hernia_lap %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.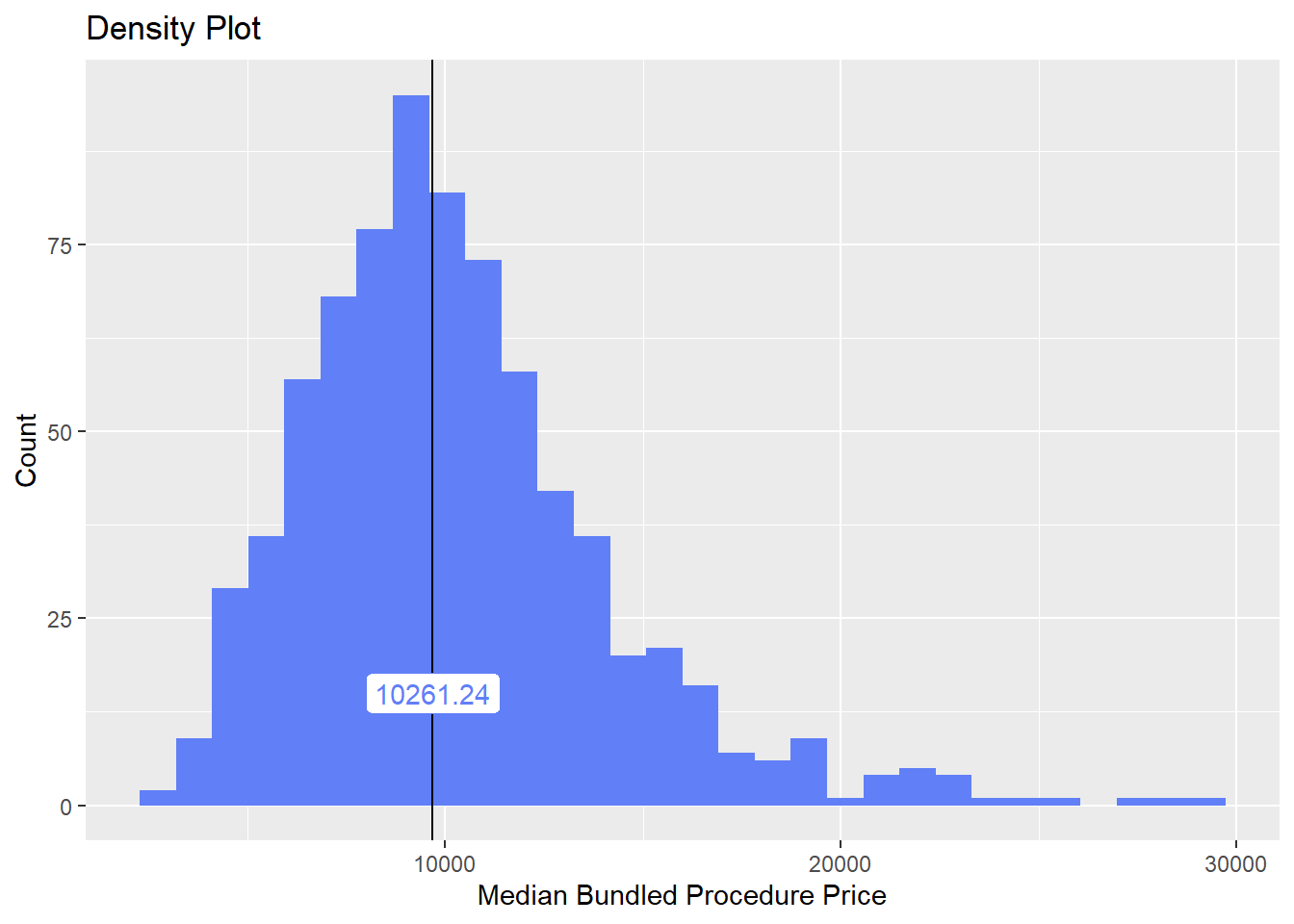
hernia_lap %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 172 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_upper 0.197
## 2 anes_bun_t_lower 0.180
## 3 path_bun_t_total 0.154
## 4 surg_bun_t_abd 0.153
## 5 path_bun_t_complete 0.131
## 6 path_bun_t_magnesium 0.124
## 7 path_bun_t_phosphorus 0.124
## 8 path_bun_t_time 0.110
## 9 surg_bun_t_mesh 0.0975
## 10 path_bun_t_metabolic 0.0956
## # ... with 162 more rowshernia_lap_btbv4 <- hernia_lap %>% btbv4() %>% .[,
primary_doctor := pmap(.l=list(doctor_npi1=doctor_npi_str1,
doctor_npi2=doctor_npi_str2,
class_reqs="Surgery"#,
# specialization_reqs = ""
),
.f=calculate_primary_doctor) %>% as.character()
] %>%
#Filter out any procedures where our doctors fail both criteria.
.[!(primary_doctor %in% c("BOTH_DOC_FAIL_CRIT", "TWO_FIT_ALL_SPECS", "NONE_FIT_SPEC_REQ"))] %>%
.[,primary_doctor_npi := fifelse(primary_doctor==doctor_str1,
doctor_npi_str1,
doctor_npi_str2)] %>%
.[,`:=`(procedure_type=6, procedure_modifier="Hernia Laparoscopy Repair")]## [1] "multiple meet class req"
## [1] "ROBERT PATTERSON" "MILDA SHAPIRO"
## [1] "multiple meet class req"
## [1] "RODRICK D MCKINLAY" "NICHOLAS J PAULK"
## [1] "multiple meet class req"
## [1] "TREVOR DAY" "CAMERON TALMAGE BLACK"
## [1] "multiple meet class req"
## [1] "MILDA SHAPIRO" "WILLIAM NOEL PEUGH"
## [1] "multiple meet class req"
## [1] "MATTHEW T BAKER" "BRIAN GILL"
## [1] "multiple meet class req"
## [1] "ROBERT PATTERSON" "WILLIAM NOEL PEUGH"
## [1] "multiple meet class req"
## [1] "KYLE G DUNNING" "MEGAN WOLTHUIS GRUNANDER"
## [1] "multiple meet class req"
## [1] "DAVID E SKARDA" "DEVAN GRINER"
## [1] "multiple meet class req"
## [1] "DANIELLE M. ADAMS" "VANESSA MARIE HART"hernia_lap_btbv4 %>% count(most_important_fac) %>% arrange(desc(n)) %>% View()herna_n_lap
hernia_n_lap %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
hernia_n_lap %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 329 x 2
## name correlation
## <chr> <dbl>
## 1 duration_mean 0.56
## 2 faci_bun_t_hospital 0.399
## 3 faci_bun_t_care 0.379
## 4 surg_bun_t_skin 0.362
## 5 surg_bun_t_graft 0.356
## 6 surg_bun_t_intestine 0.348
## 7 surg_bun_t_small 0.348
## 8 surg_bun_t_musc 0.340
## 9 faci_bun_t_initial 0.337
## 10 medi_bun_t_inject 0.333
## # ... with 319 more rowshernia_n_lap %>% get_tag_density_information("faci_bun_t_hospital") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ faci_bun_t_hospital"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 2530## $dist_plots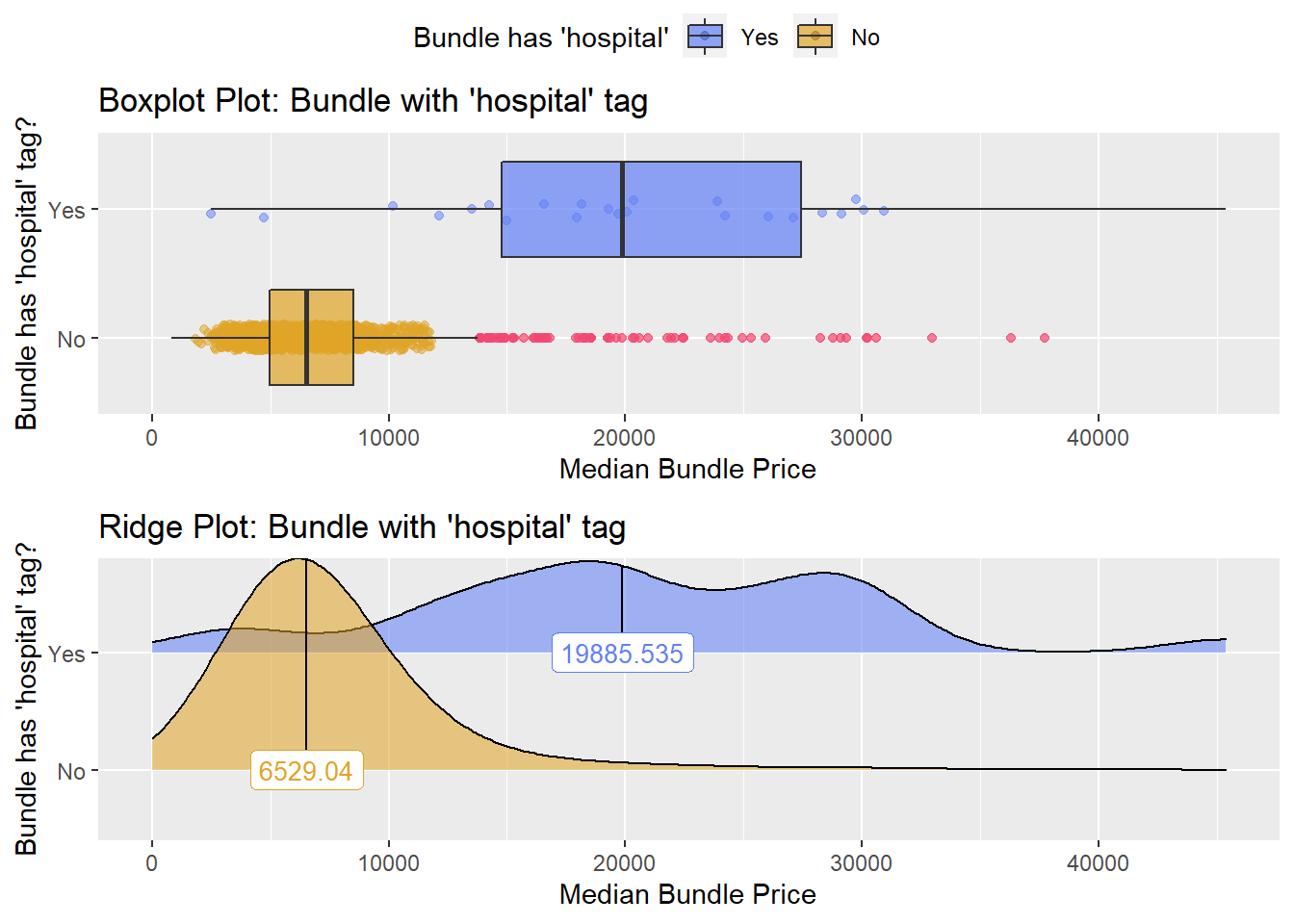
##
## $stat_tables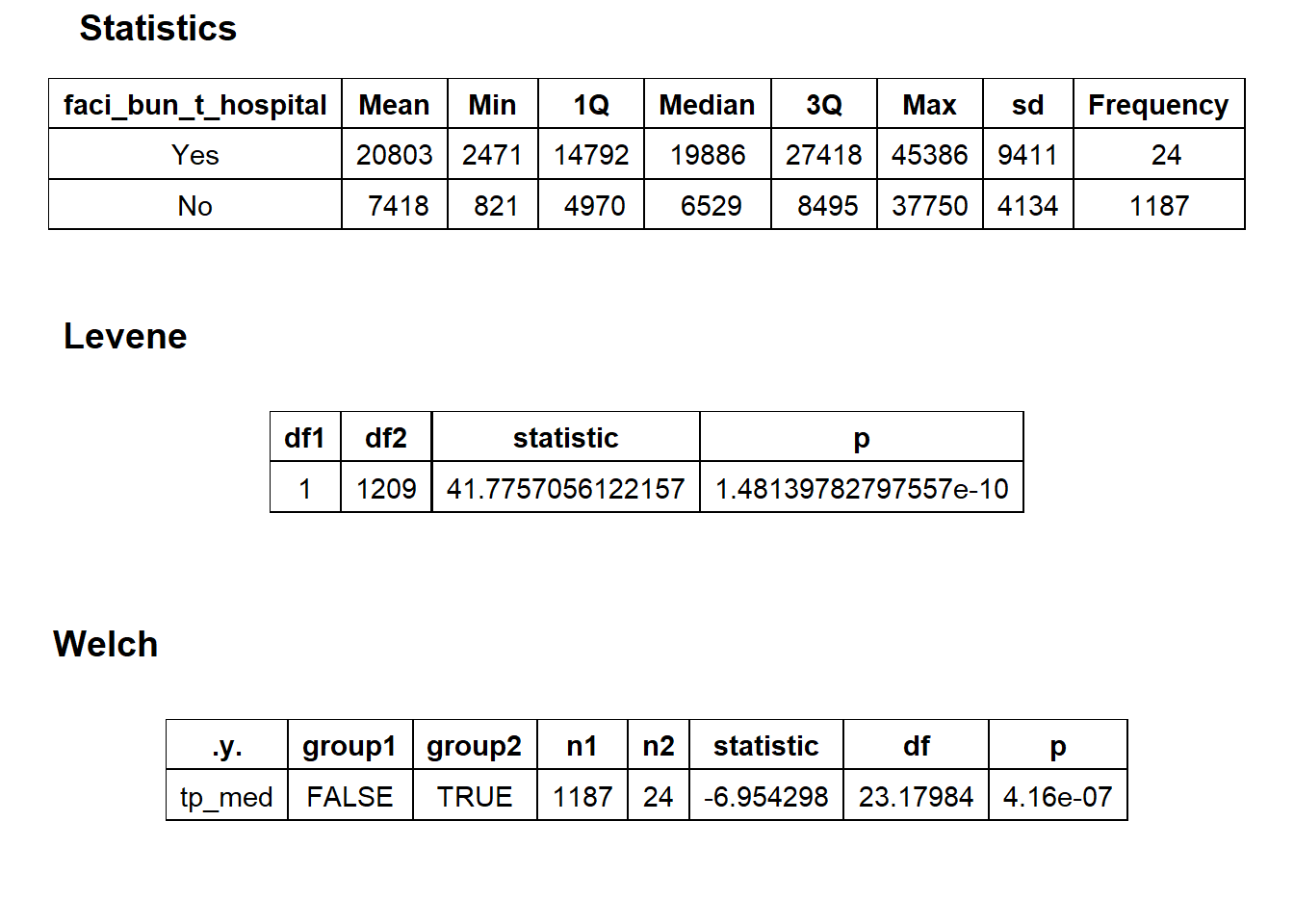
hernia_n_lap <- hernia_n_lap[faci_bun_t_hospital==F]hernia_n_lap %>% get_tag_density_information("surg_bun_t_skin") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_skin"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 1940## $dist_plots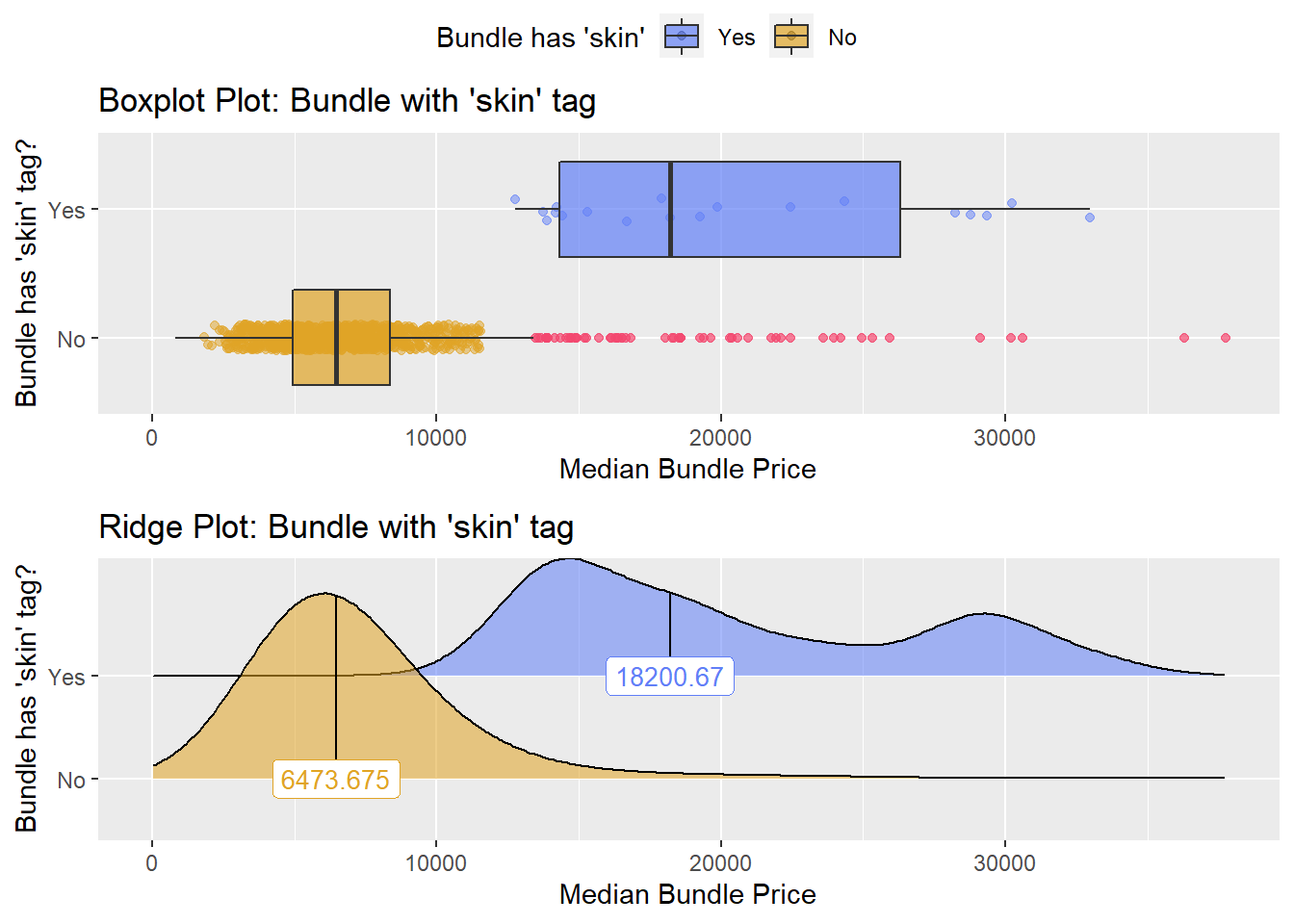
##
## $stat_tables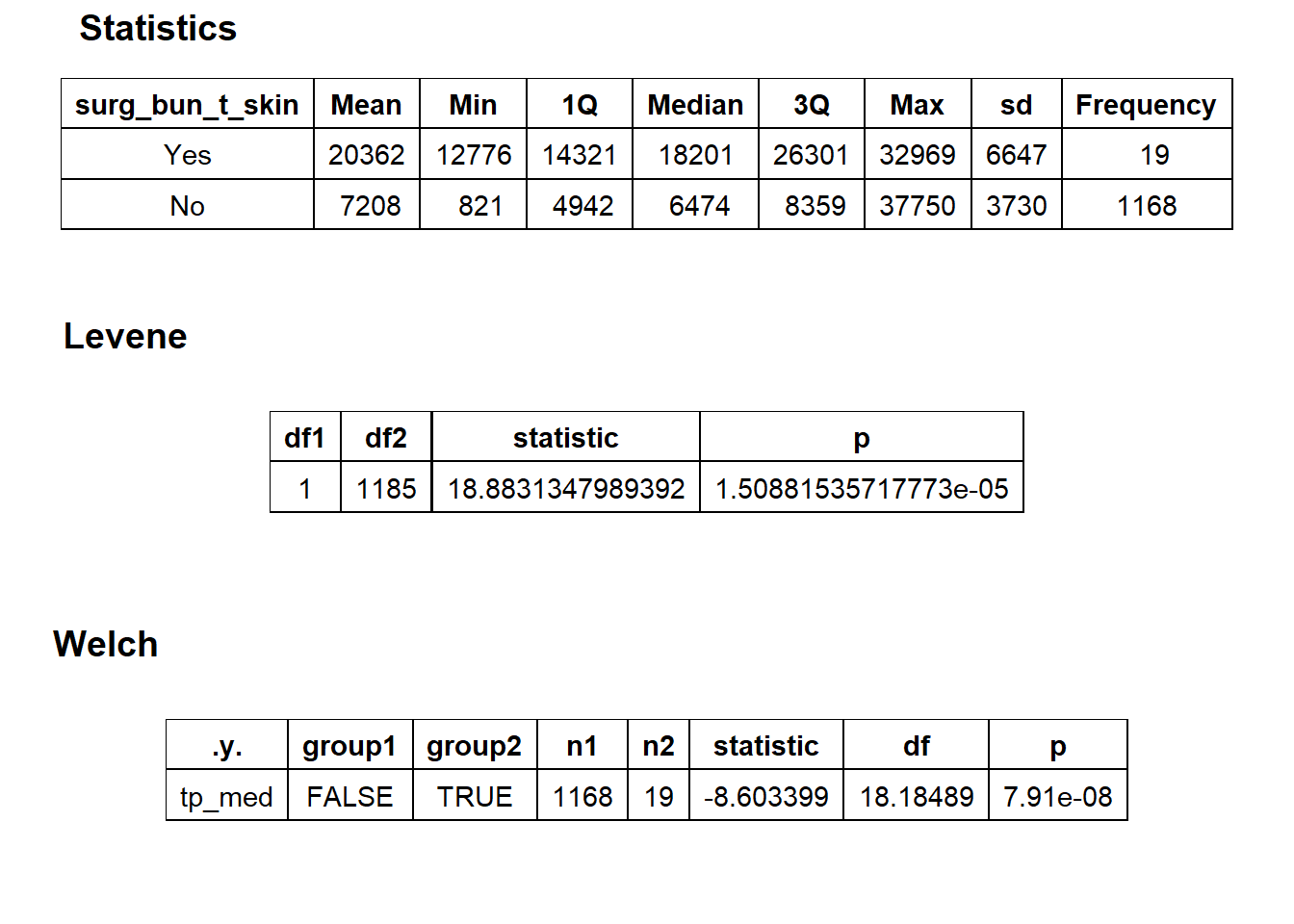
hernia_n_lap <- hernia_n_lap[surg_bun_t_skin == F & surg_bun_t_graft==F ]hernia_n_lap %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.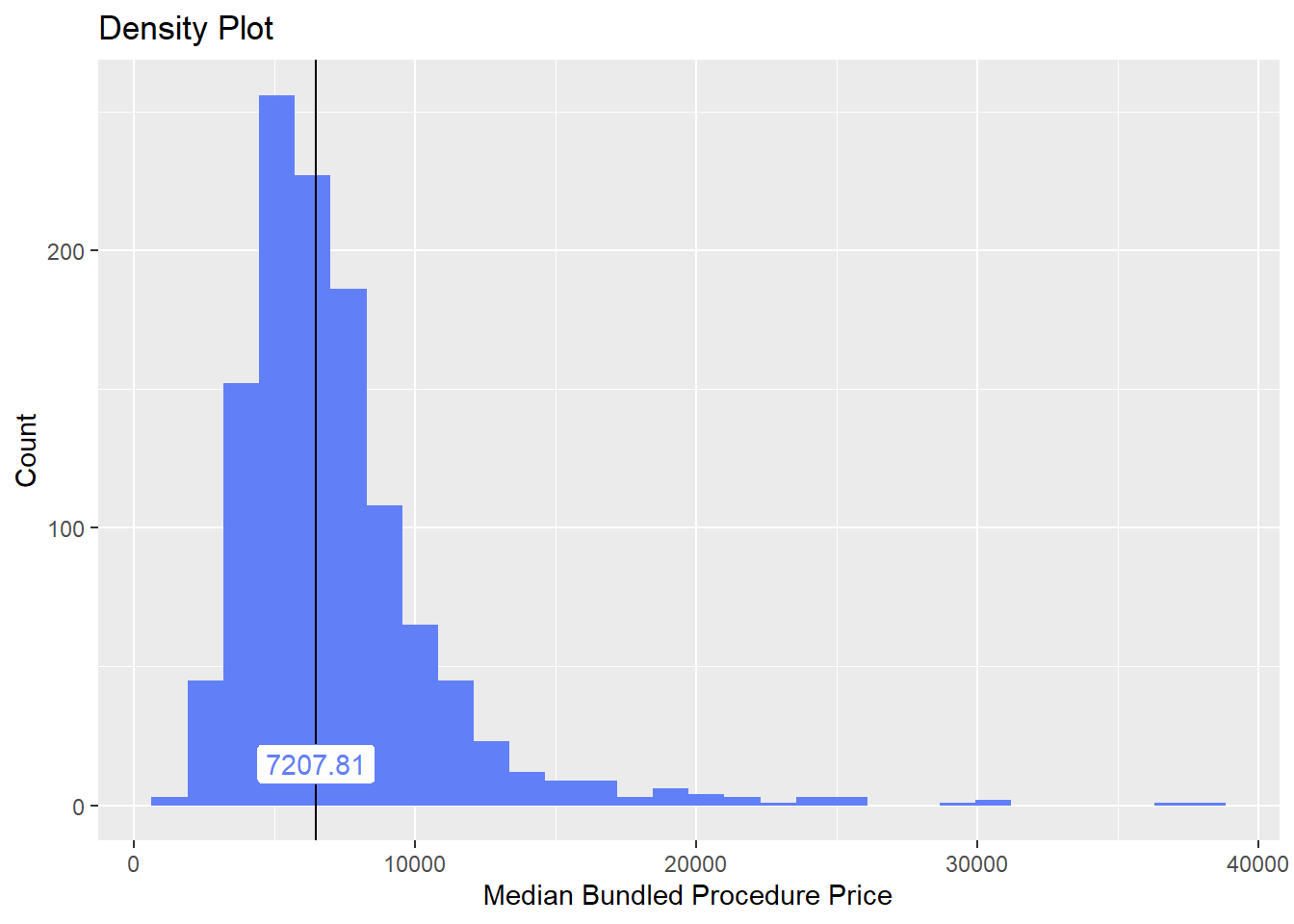
hernia_n_lap %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 301 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.504
## 2 duration_mean 0.491
## 3 surg_bun_t_esoph 0.331
## 4 radi_bun_t_ct 0.279
## 5 radi_bun_t_chest 0.272
## 6 surg_bun_t_hernia 0.268
## 7 surg_bun_t_repair 0.257
## 8 radi_bun_t_upper 0.240
## 9 radi_bun_t_contrast 0.239
## 10 anes_bun_t_upper 0.237
## # ... with 291 more rowshernia_n_lap %>% get_tag_density_information("surg_bun_t_esoph") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_esoph"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 492## $dist_plots
##
## $stat_tables
hernia_n_lap <- hernia_n_lap[surg_bun_t_esoph == F]hernia_n_lap %>% get_tag_density_information("radi_bun_t_chest") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ radi_bun_t_chest"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 2480## $dist_plots
##
## $stat_tables
hernia_n_lap <- hernia_n_lap[radi_bun_t_chest == F]hernia_n_lap %>% get_tag_density_information("radi_bun_t_contrast") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ radi_bun_t_contrast"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 2080## $dist_plots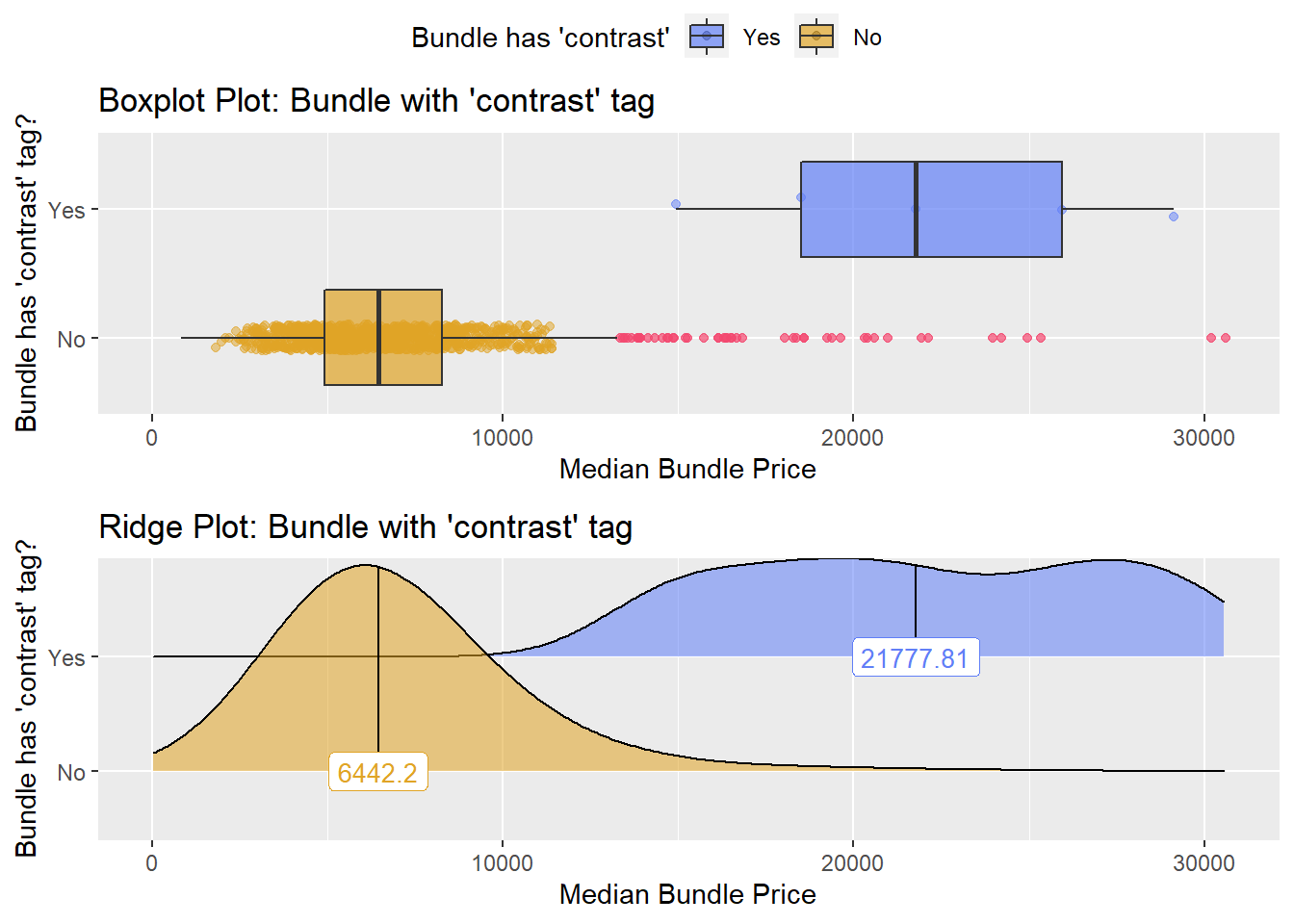
##
## $stat_tables
hernia_n_lap <- hernia_n_lap[radi_bun_t_contrast == F]hernia_n_lap %>% get_tag_density_information("surg_bun_t_umbil") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_umbil"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 595## $dist_plots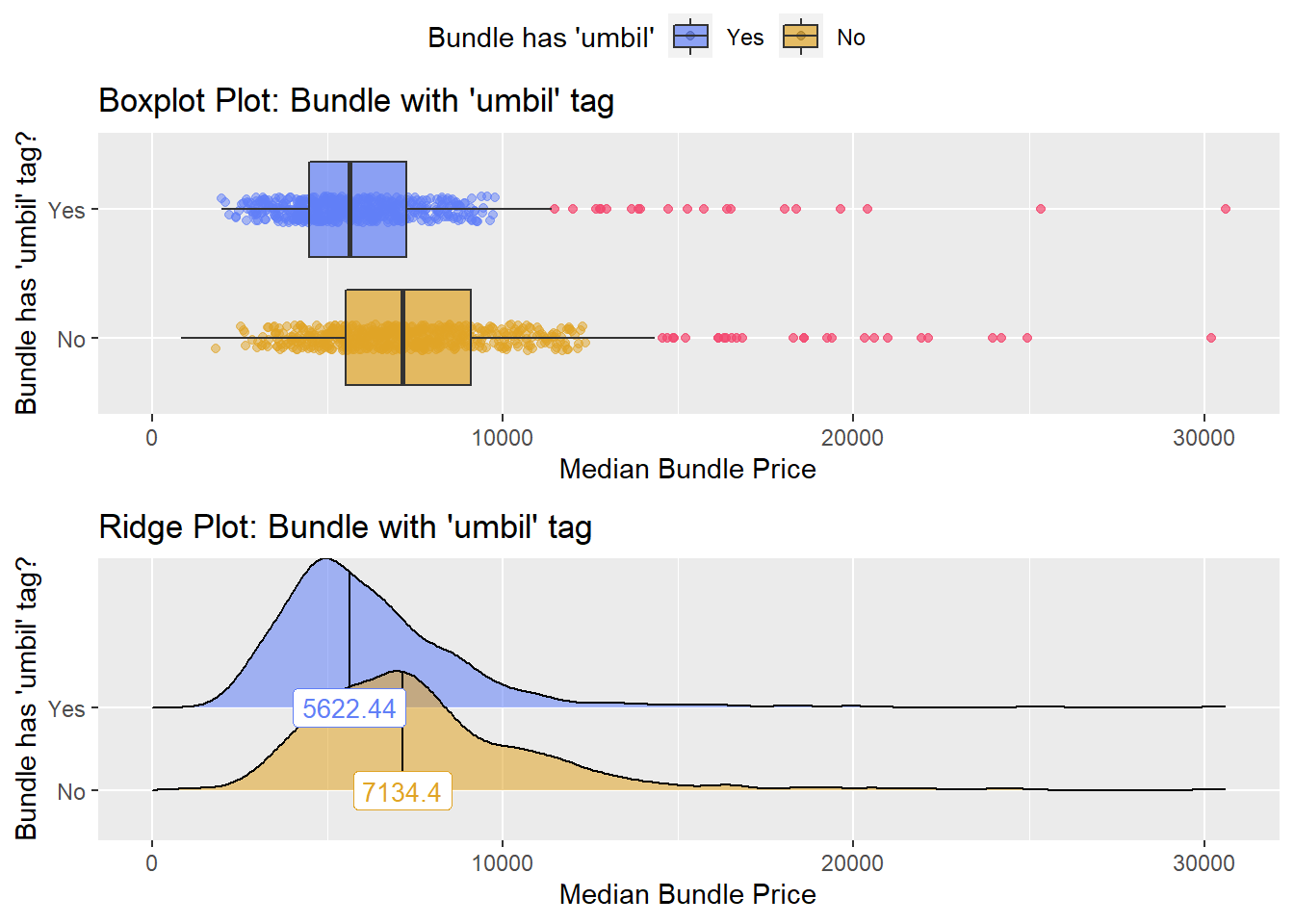
##
## $stat_tables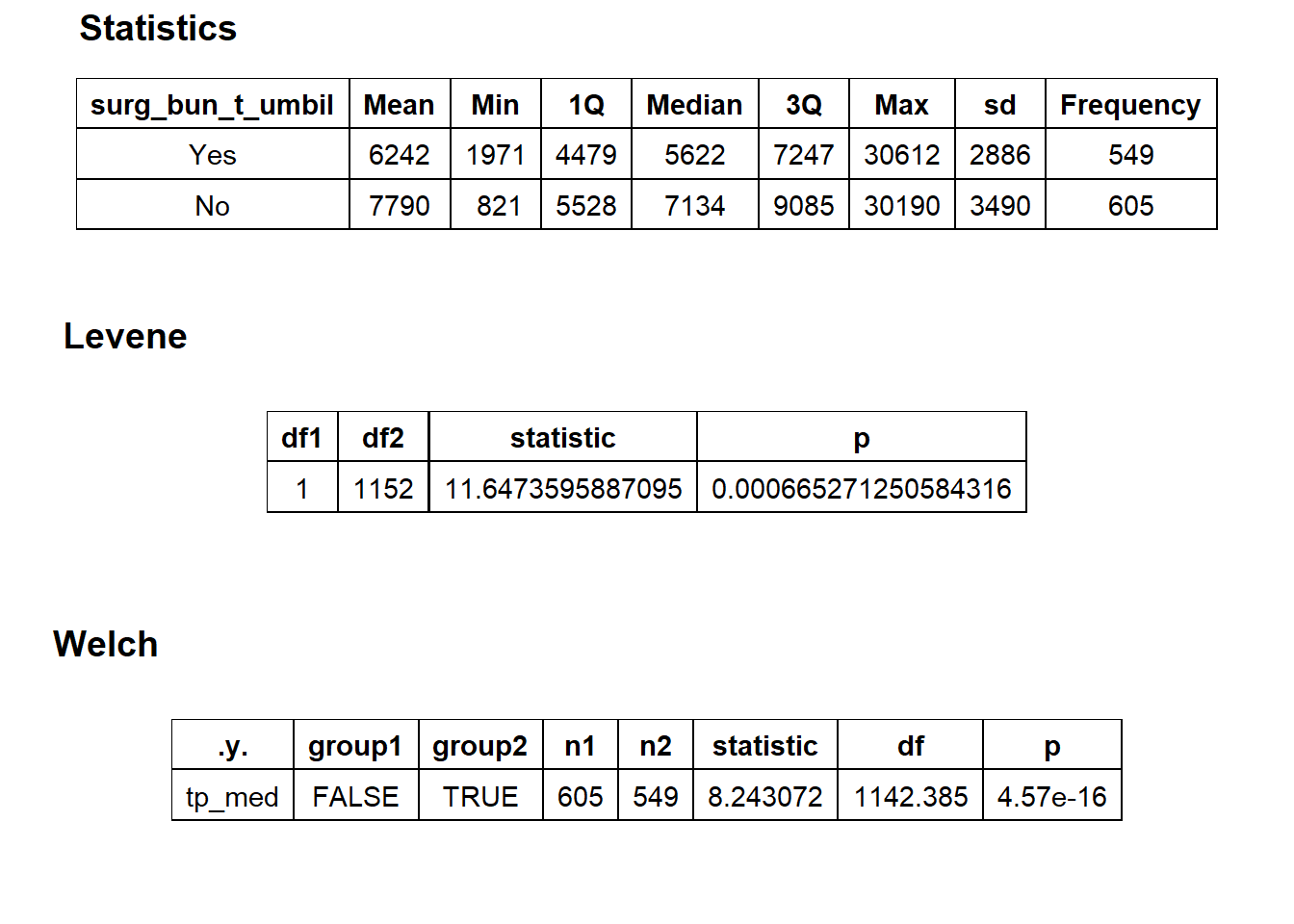
hernia_n_lap %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.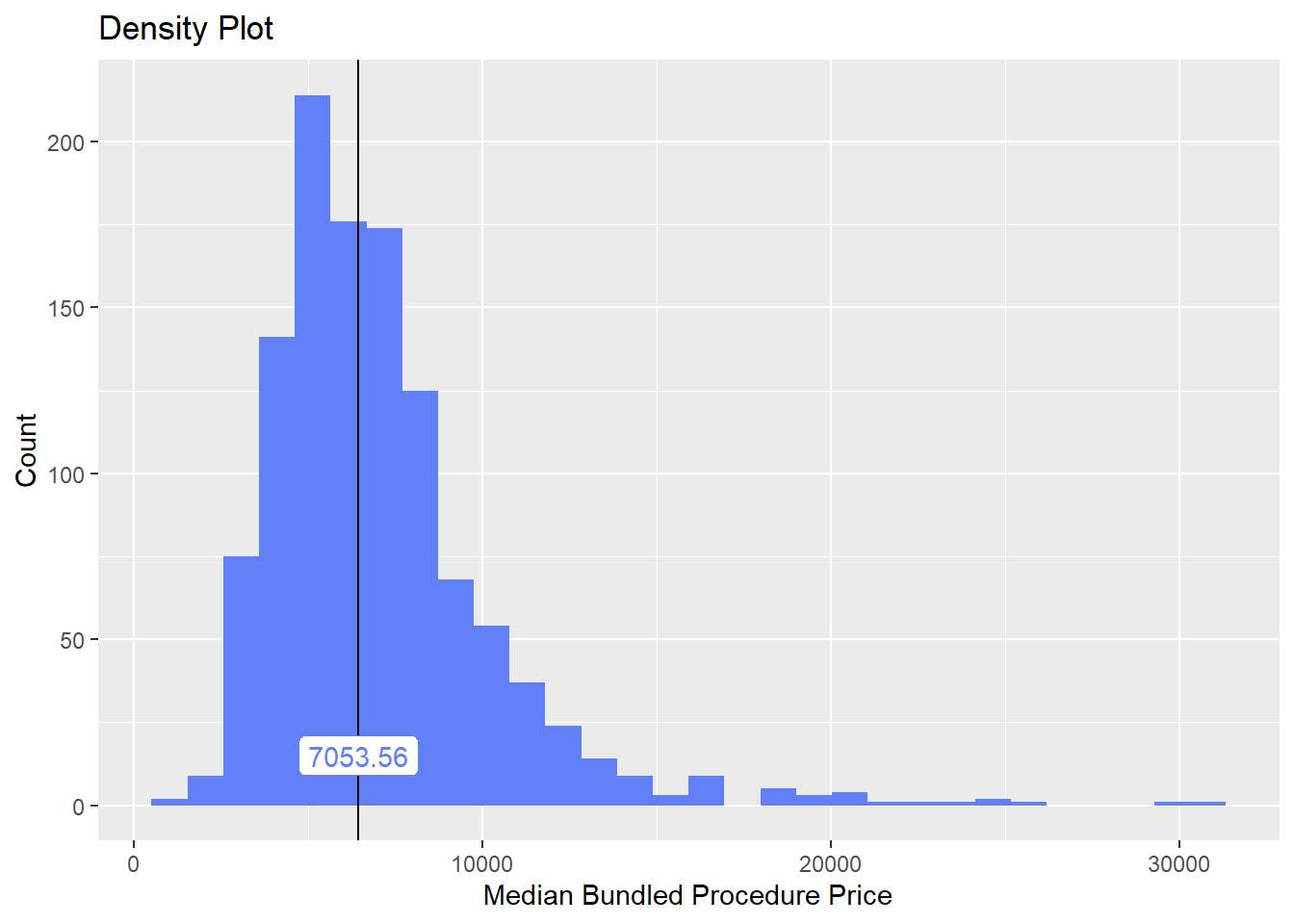
hernia_n_lap %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 290 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.356
## 2 duration_mean 0.343
## 3 surg_bun_t_hernia 0.296
## 4 surg_bun_t_repair 0.270
## 5 surg_bun_t_mesh 0.250
## 6 surg_bun_t_injection 0.236
## 7 surg_bun_t_umbil 0.234
## 8 surg_bun_t_art 0.210
## 9 surg_bun_t_arthro 0.210
## 10 surg_bun_t_arthroscopy 0.210
## # ... with 280 more rowshernia_n_lap[surg_bun_t_arthroscopy==T] %>% nrow() #%>% get_tag_density_information("surg_bun_t_arthroscopy") %>% print()## [1] 1hernia_n_lap[surg_bun_t_uterus==T] %>% nrow()## [1] 3hernia_n_lap[path_bun_t_typing==T] %>% nrow()## [1] 7hernia_n_lap[path_bun_t_anti==T] %>% nrow()## [1] 5hernia_n_lap[path_bun_t_tissue==T] %>% nrow()## [1] 98hernia_n_lap[surg_bun_t_interlaminar==T] %>% nrow()## [1] 7hernia_n_lap[faci_bun_t_est==T] %>% nrow()## [1] 53hernia_n_lap[path_bun_t_urinalysis==T] %>% nrow()## [1] 8hernia_n_lap <- hernia_n_lap[surg_bun_t_arthroscopy==F & surg_bun_t_uterus==F & path_bun_t_typing==F & path_bun_t_anti==F & surg_bun_t_interlaminar==F & path_bun_t_urinalysis==F]hernia_n_lap %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
hernia_n_lap %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 258 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.366
## 2 duration_mean 0.356
## 3 surg_bun_t_hernia 0.307
## 4 surg_bun_t_repair 0.294
## 5 surg_bun_t_umbil 0.271
## 6 surg_bun_t_mesh 0.268
## 7 surg_bun_t_injection 0.232
## 8 surg_bun_t_tum 0.187
## 9 path_bun_t_patho 0.148
## 10 path_bun_t_pathologist 0.148
## # ... with 248 more rowshernia_n_lap %>% get_tag_density_information("path_bun_t_patho") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ path_bun_t_patho"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 914## $dist_plots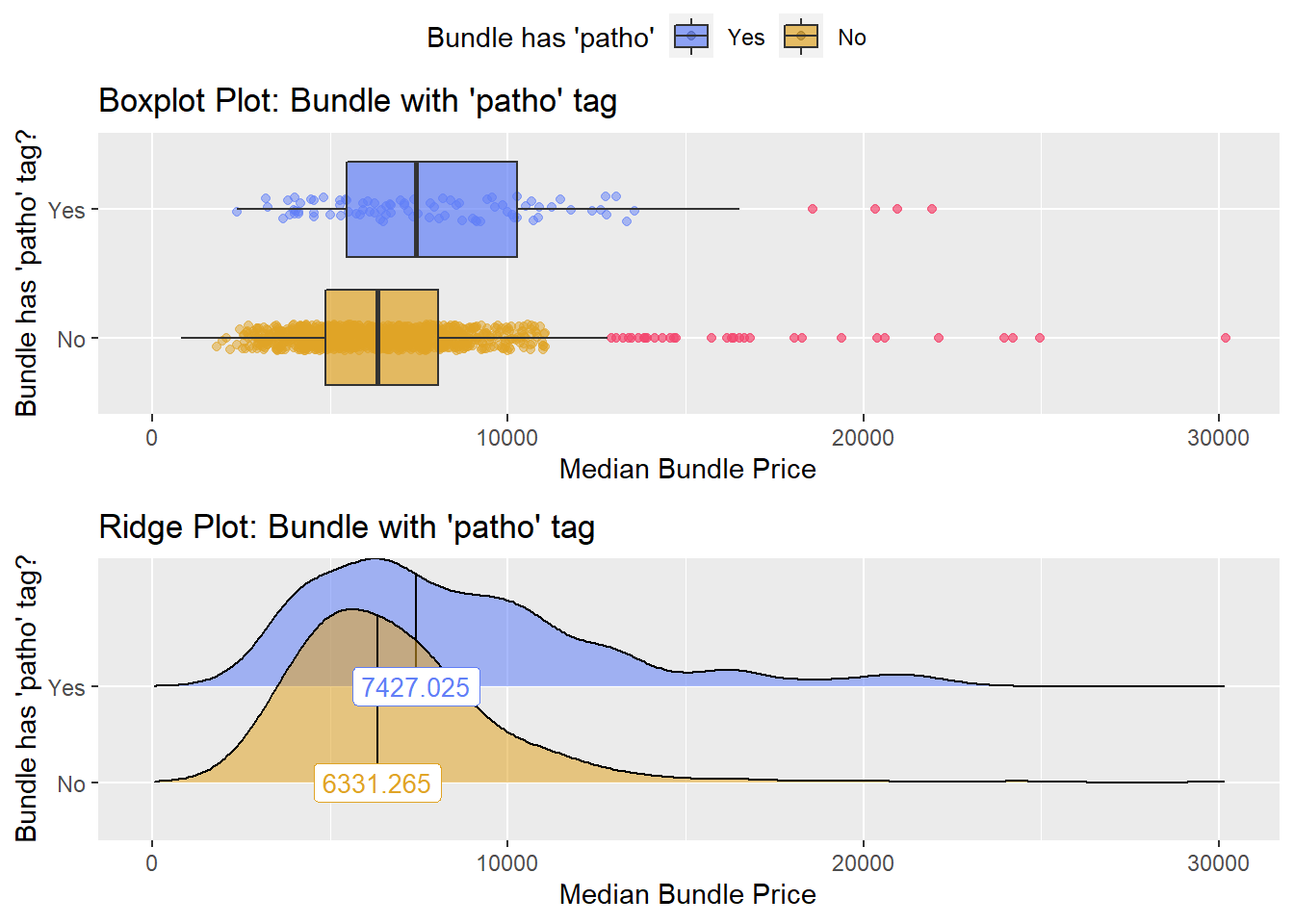
##
## $stat_tables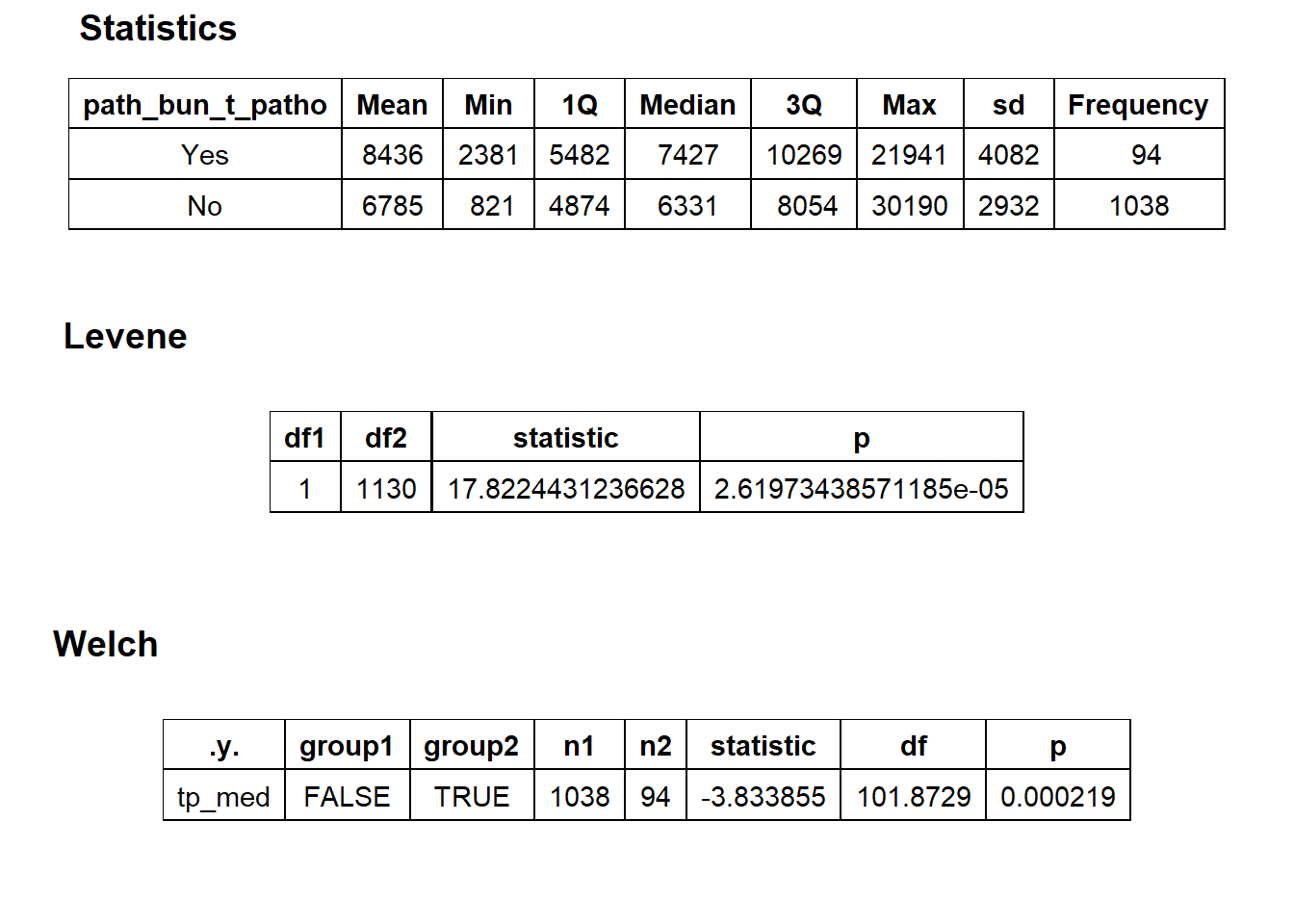
hernia_n_lap <- hernia_n_lap[path_bun_t_patho==F]hernia_n_lap %>% get_tag_density_information("medi_bun_t_normal") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_normal"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 2150## $dist_plots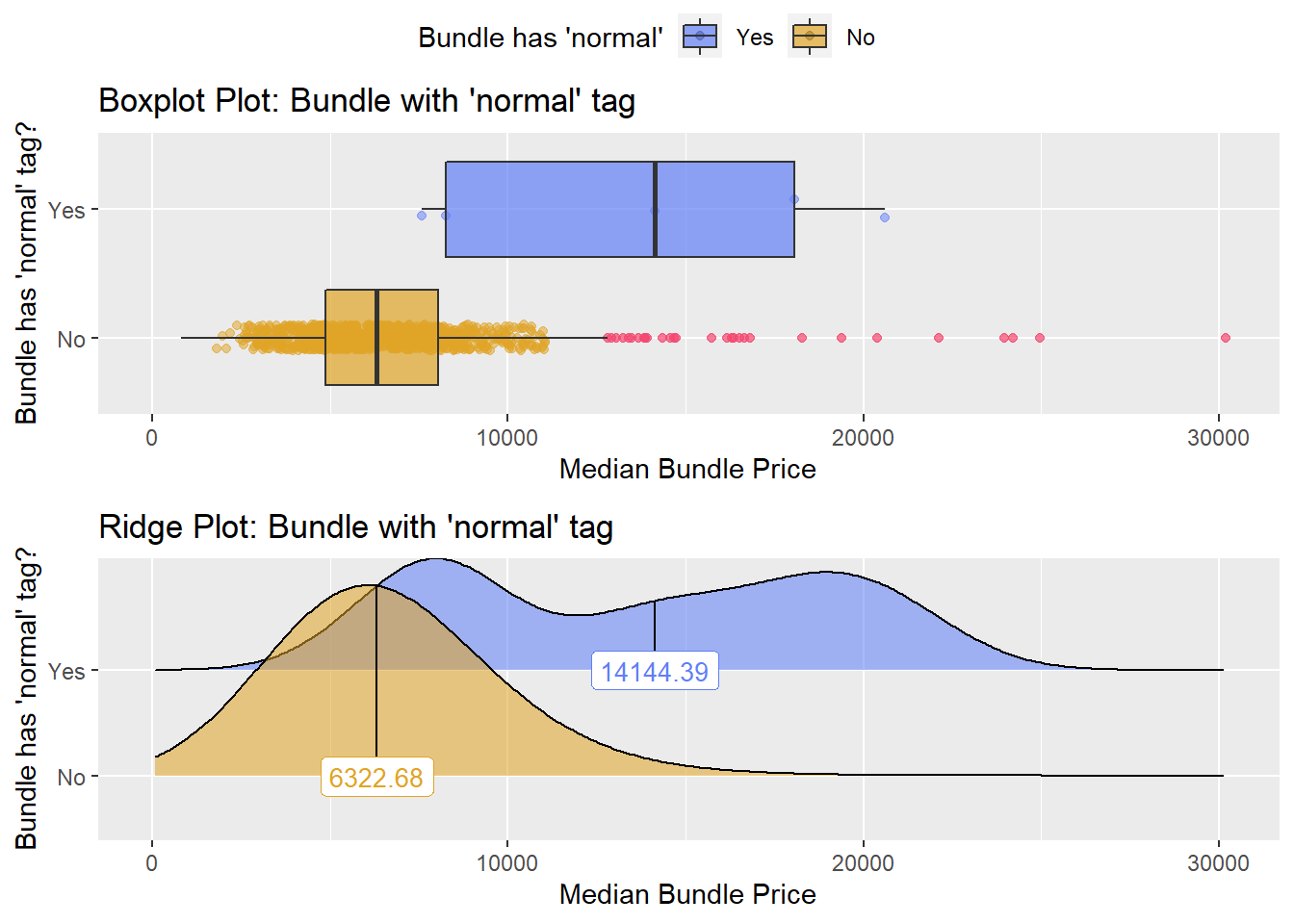
##
## $stat_tables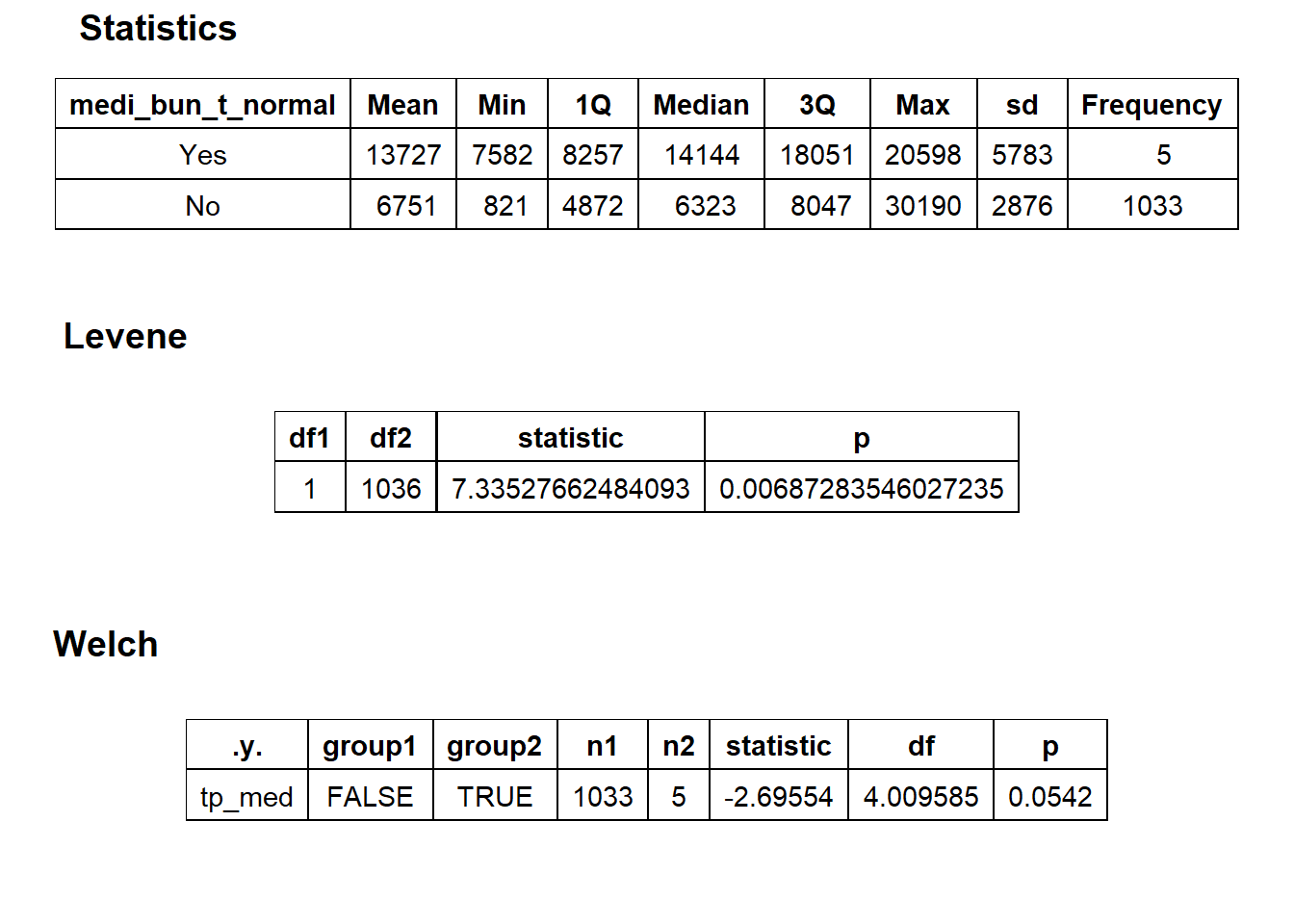
hernia_n_lap <- hernia_n_lap[medi_bun_t_normal==F]hernia_n_lap %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.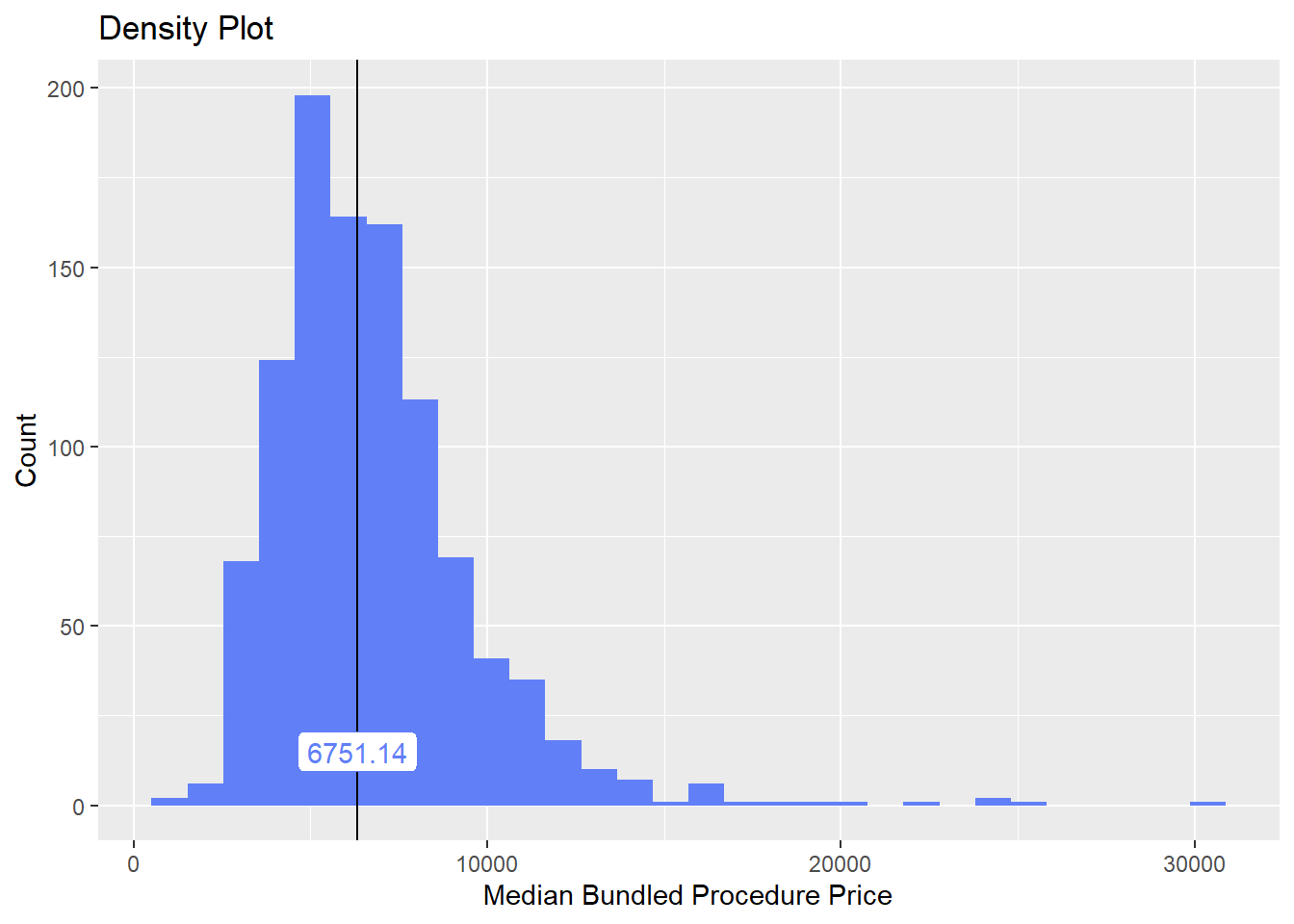
hernia_n_lap %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 207 x 2
## name correlation
## <chr> <dbl>
## 1 duration_max 0.318
## 2 surg_bun_t_hernia 0.314
## 3 duration_mean 0.310
## 4 surg_bun_t_repair 0.276
## 5 surg_bun_t_mesh 0.259
## 6 surg_bun_t_umbil 0.258
## 7 surg_bun_t_injection 0.212
## 8 medi_bun_t_splt 0.166
## 9 surg_bun_t_revise 0.152
## 10 surg_bun_t_ablate 0.148
## # ... with 197 more rowshernia_n_lap[surg_bun_t_tumor==T] %>% nrow()## [1] 0hernia_n_lap[surg_bun_t_injection==T] %>% nrow()## [1] 80hernia_n_lap[medi_bun_t_splt==T] %>% nrow()## [1] 1hernia_n_lap[surg_bun_t_revise==T] %>% nrow()## [1] 3hernia_n_lap[surg_bun_t_ablate==T] %>% nrow()## [1] 1hernia_n_lap[surg_bun_t_ear==T] %>% nrow()## [1] 1hernia_n_lap[surg_bun_t_defect==T] %>% nrow()## [1] 1hernia_n_lap[anes_bun_t_abdom==T] %>% nrow()## [1] 60hernia_n_lap %>% get_tag_density_information("surg_bun_t_injection") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_injection"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 619## $dist_plots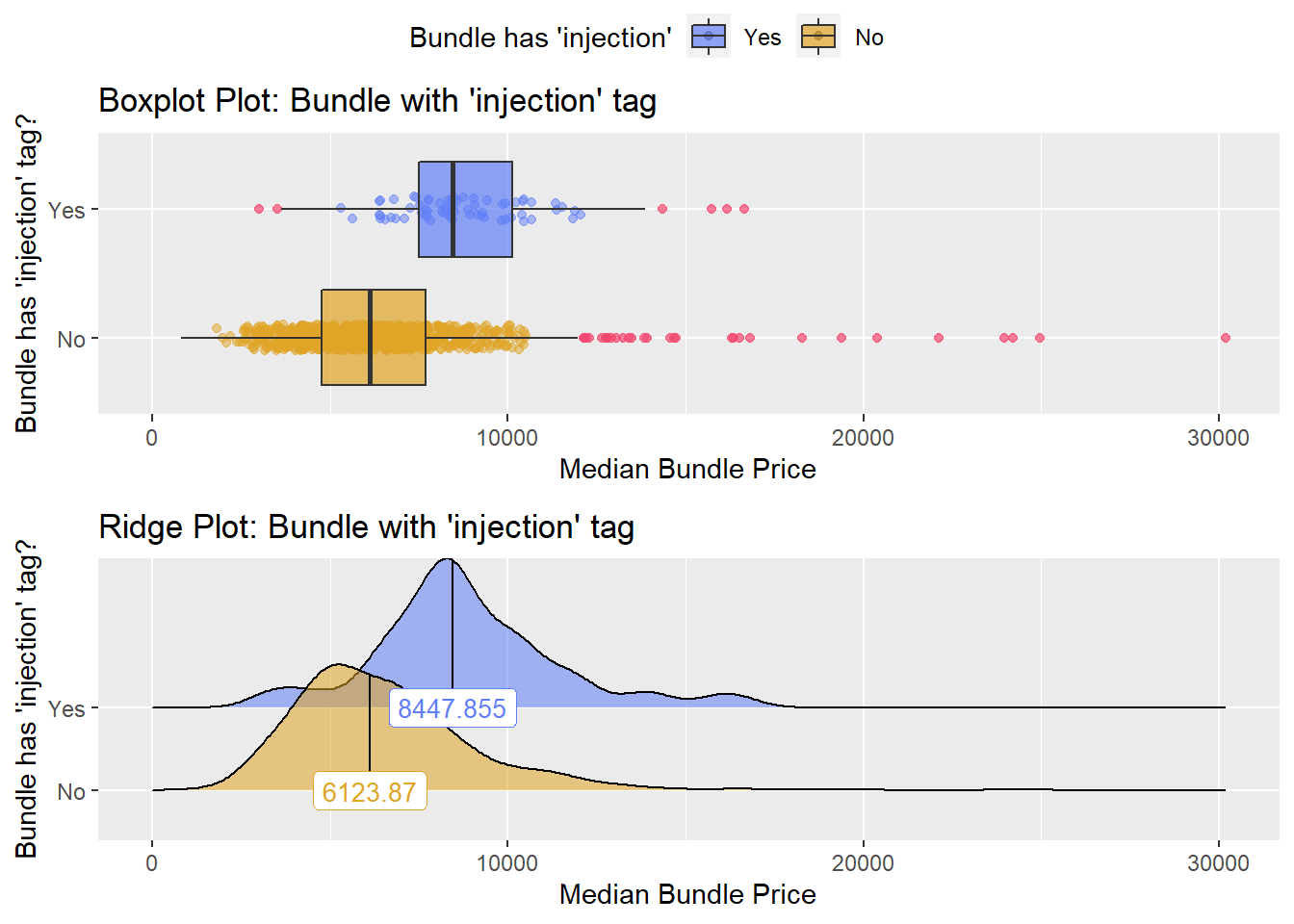
##
## $stat_tables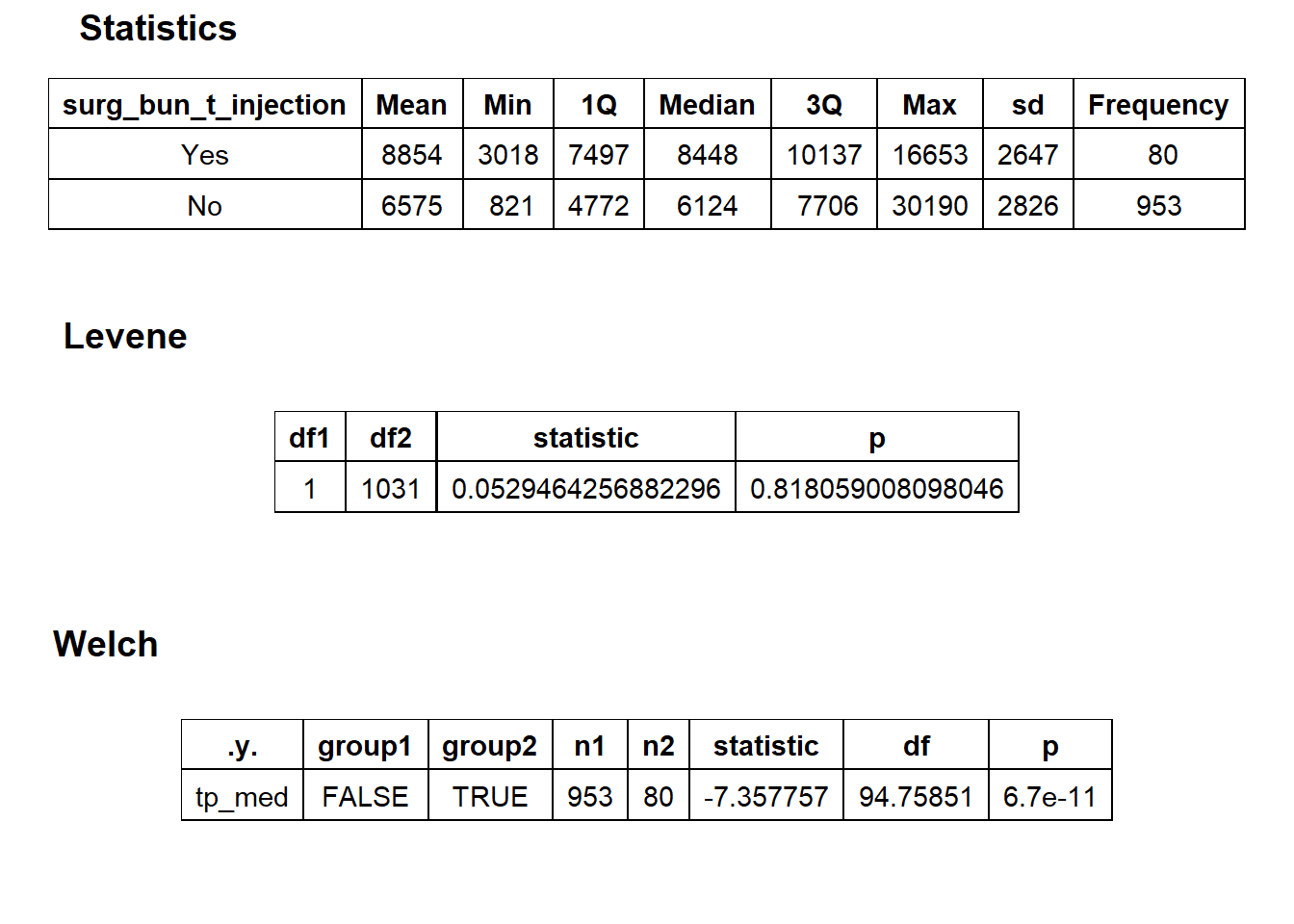
hernia_n_lap %>% get_tag_density_information("anes_bun_t_abdom") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ anes_bun_t_abdom"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 685## $dist_plots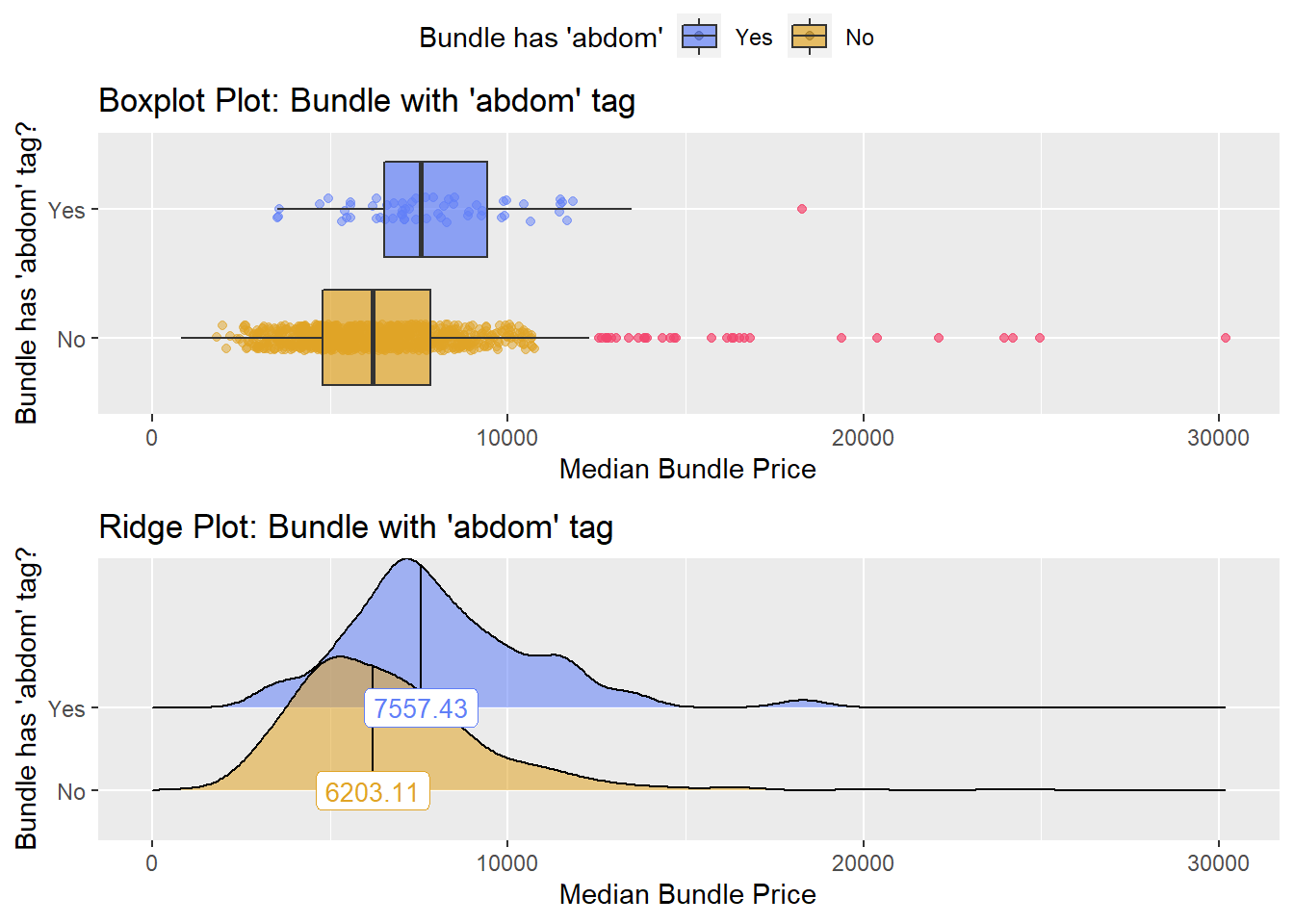
##
## $stat_tables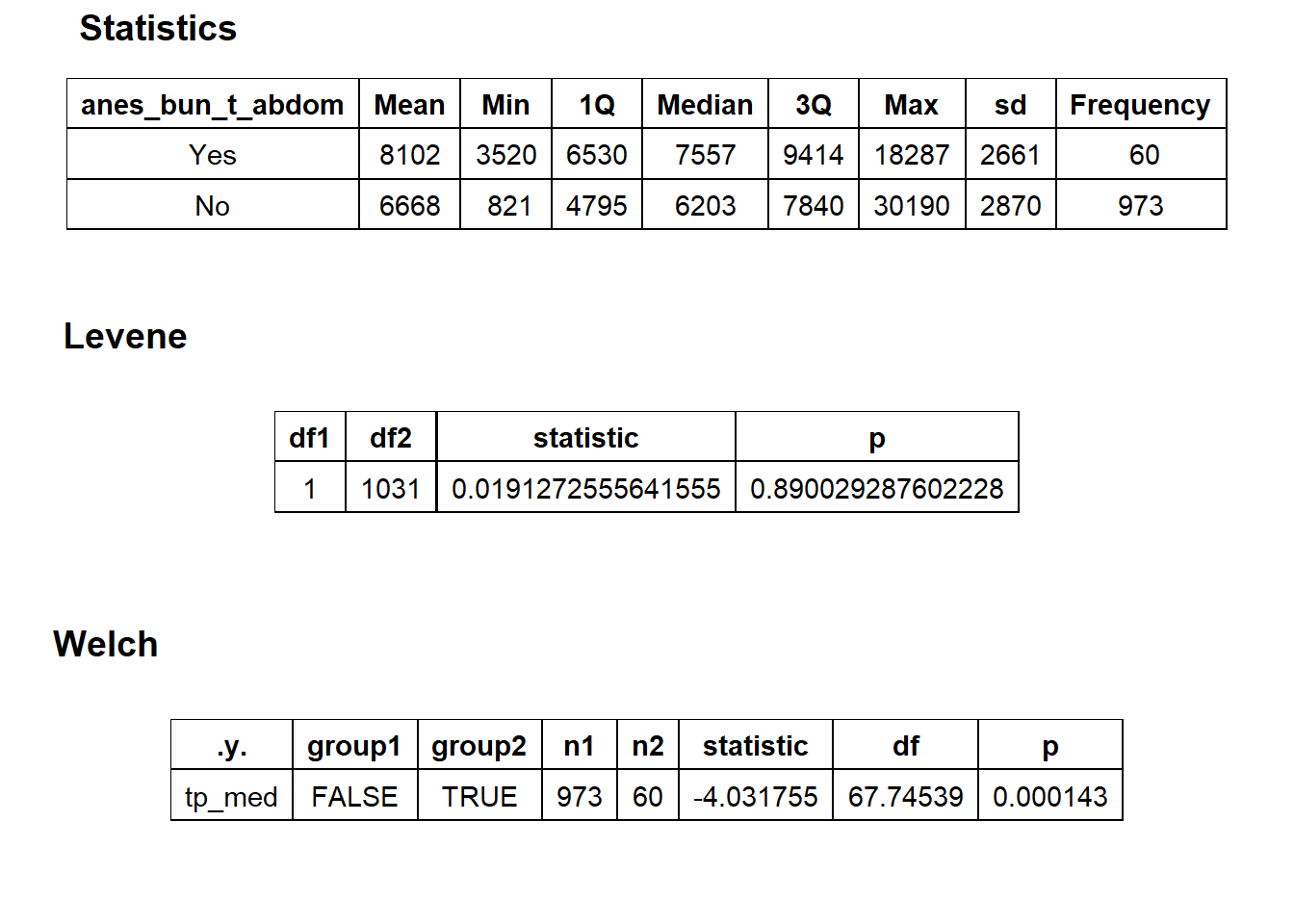
hernia_n_lap <- hernia_n_lap[surg_bun_t_injection==F & medi_bun_t_splt==F & surg_bun_t_revise ==F & surg_bun_t_ablate==F & surg_bun_t_ear==F & surg_bun_t_defect==F & anes_bun_t_abdom==F]hernia_n_lap %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
hernia_n_lap %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 173 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_hernia 0.299
## 2 surg_bun_t_repair 0.284
## 3 surg_bun_t_mesh 0.267
## 4 surg_bun_t_umbil 0.261
## 5 duration_max 0.240
## 6 duration_mean 0.229
## 7 medi_bun_t_oral 0.110
## 8 surg_bun_t_suspension 0.108
## 9 surg_bun_t_testis 0.108
## 10 anes_bun_t_anes 0.0941
## # ... with 163 more rowshernia_n_lap <- hernia_n_lap[tp_med > 2000 & tp_med < 20000]hernia_n_lap %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.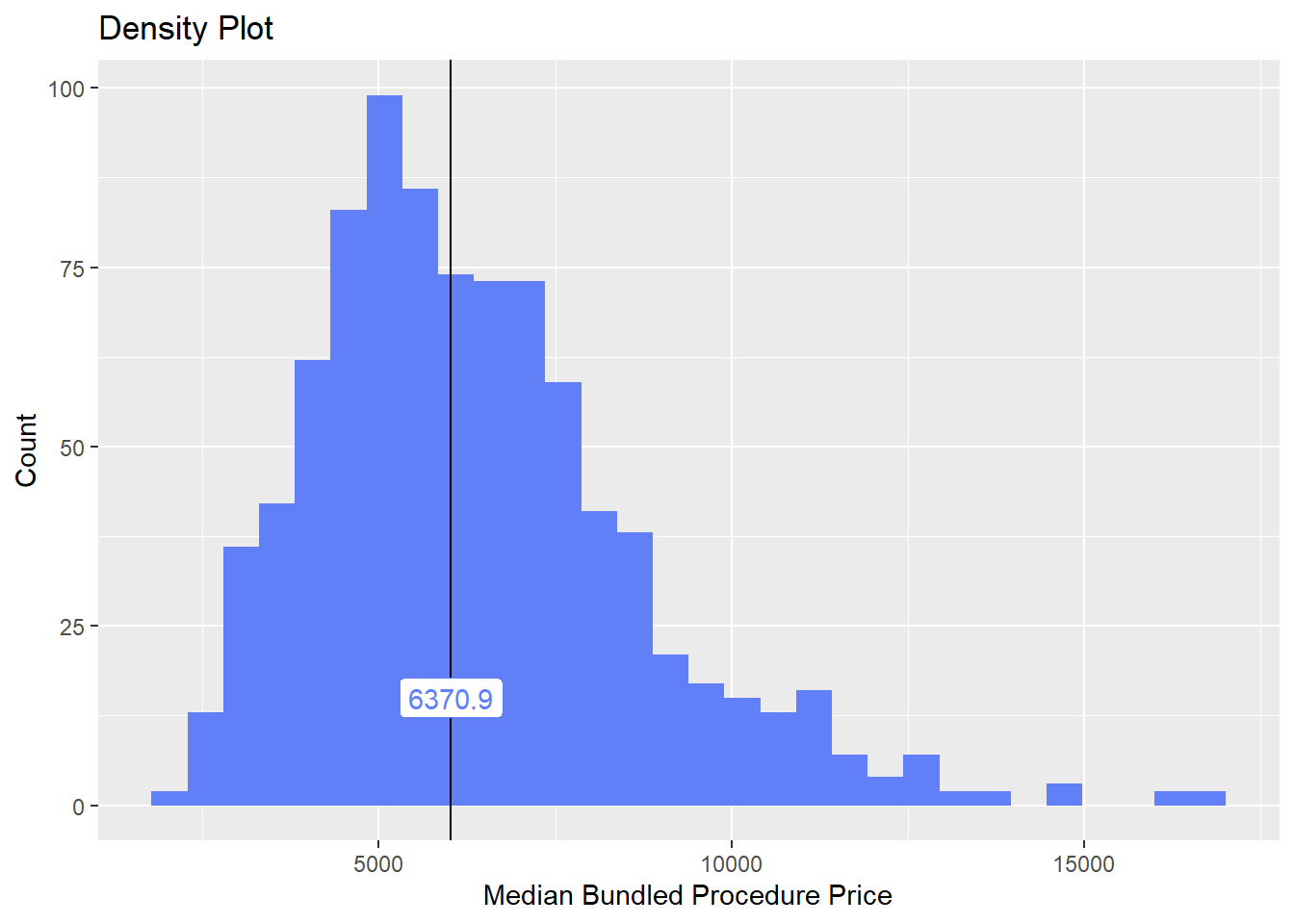
hernia_n_lap %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 172 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_hernia 0.294
## 2 surg_bun_t_umbil 0.271
## 3 surg_bun_t_repair 0.268
## 4 surg_bun_t_mesh 0.238
## 5 surg_bun_t_testis 0.129
## 6 surg_bun_t_suspension 0.129
## 7 medi_bun_t_oral 0.121
## 8 duration_max 0.119
## 9 surg_bun_t_arm 0.0949
## 10 surg_bun_t_elbow 0.0949
## # ... with 162 more rowshernia_n_lap %>% get_tag_density_information("surg_bun_t_testis") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_testis"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 670## $dist_plots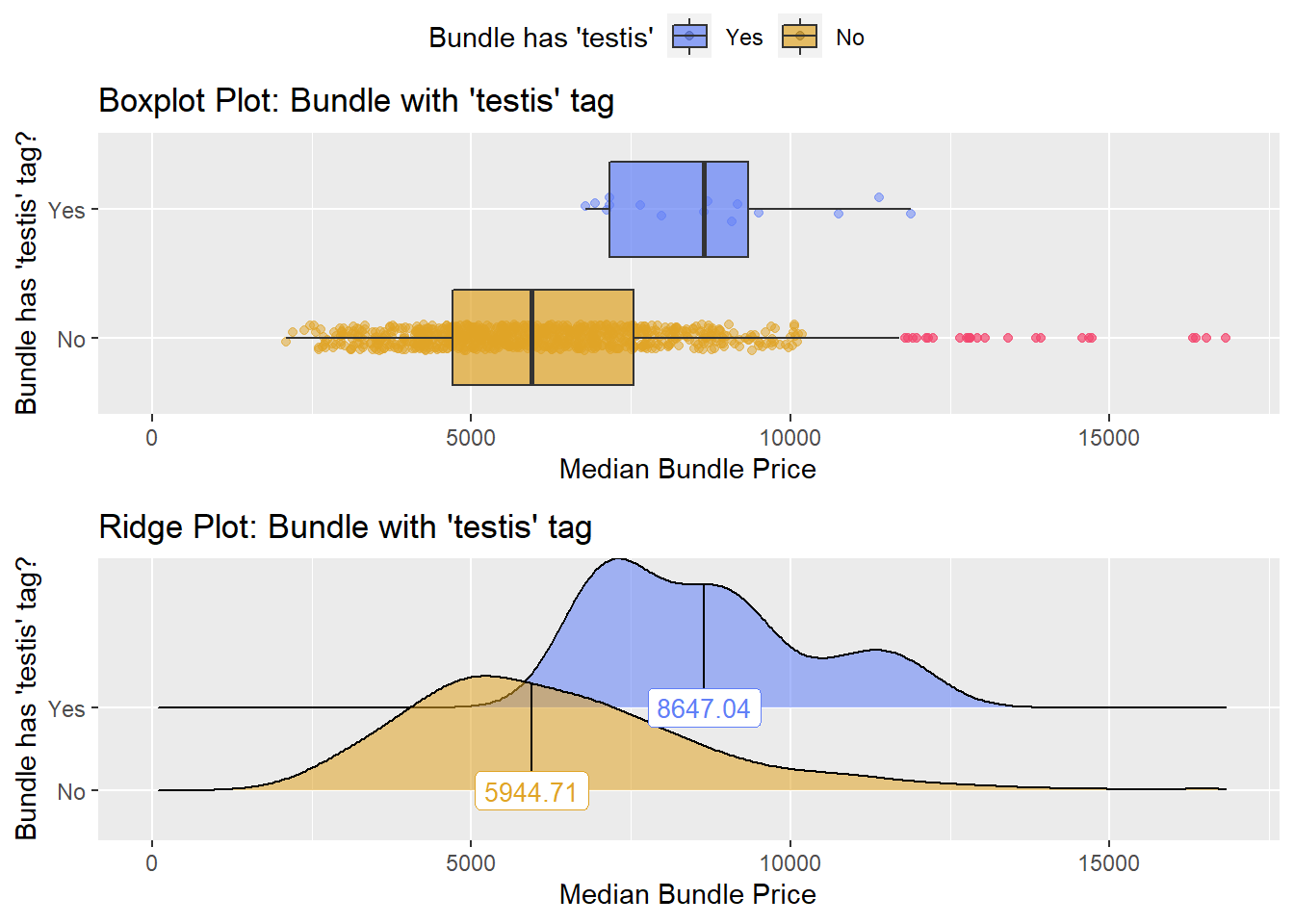
##
## $stat_tables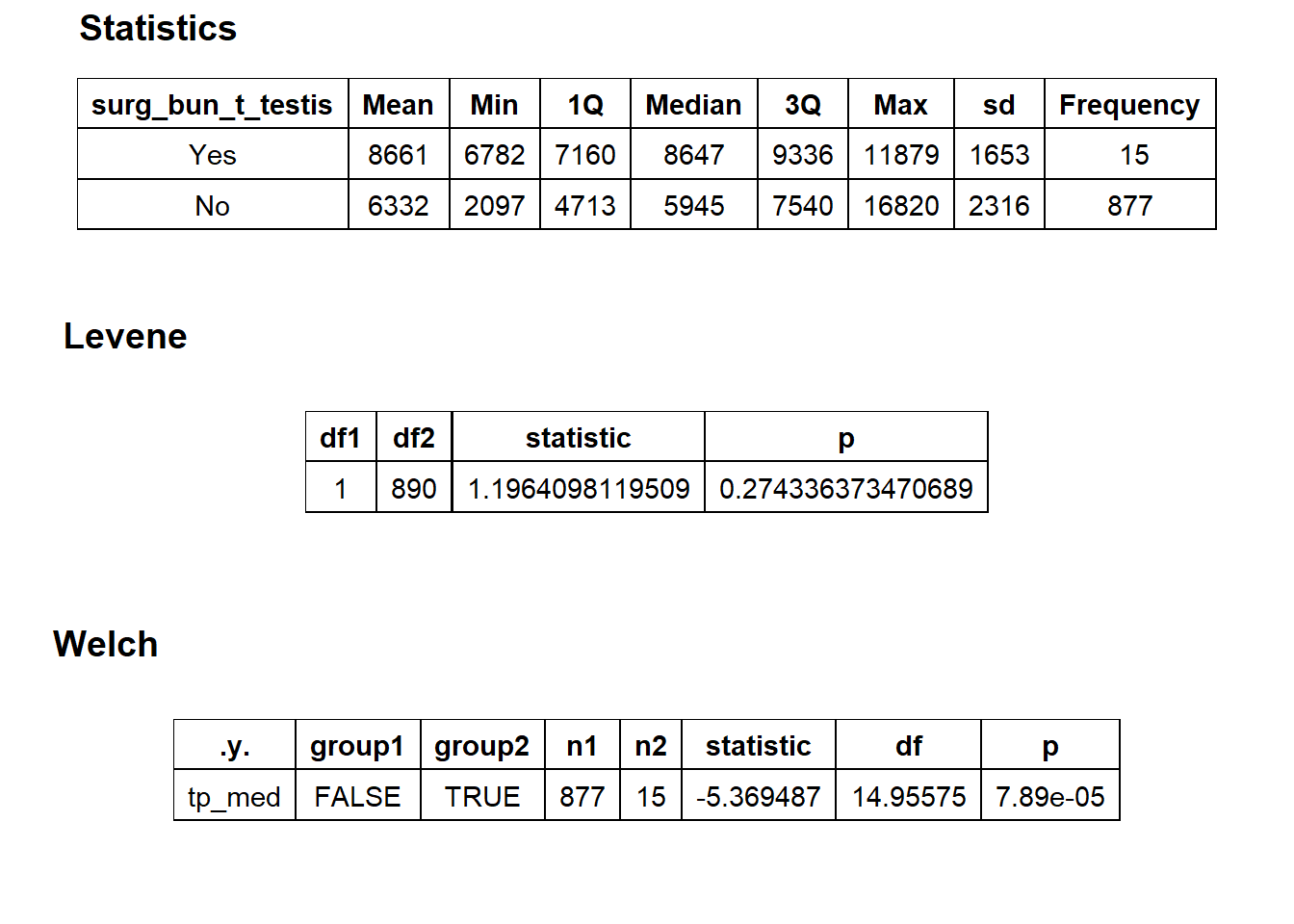
hernia_n_lap <- hernia_n_lap[surg_bun_t_testis == F ]hernia_n_lap %>% get_tag_density_information("surg_bun_t_suspension") %>% print()hernia_n_lap <- hernia_n_lap[surg_bun_t_suspension == F ]hernia_n_lap %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.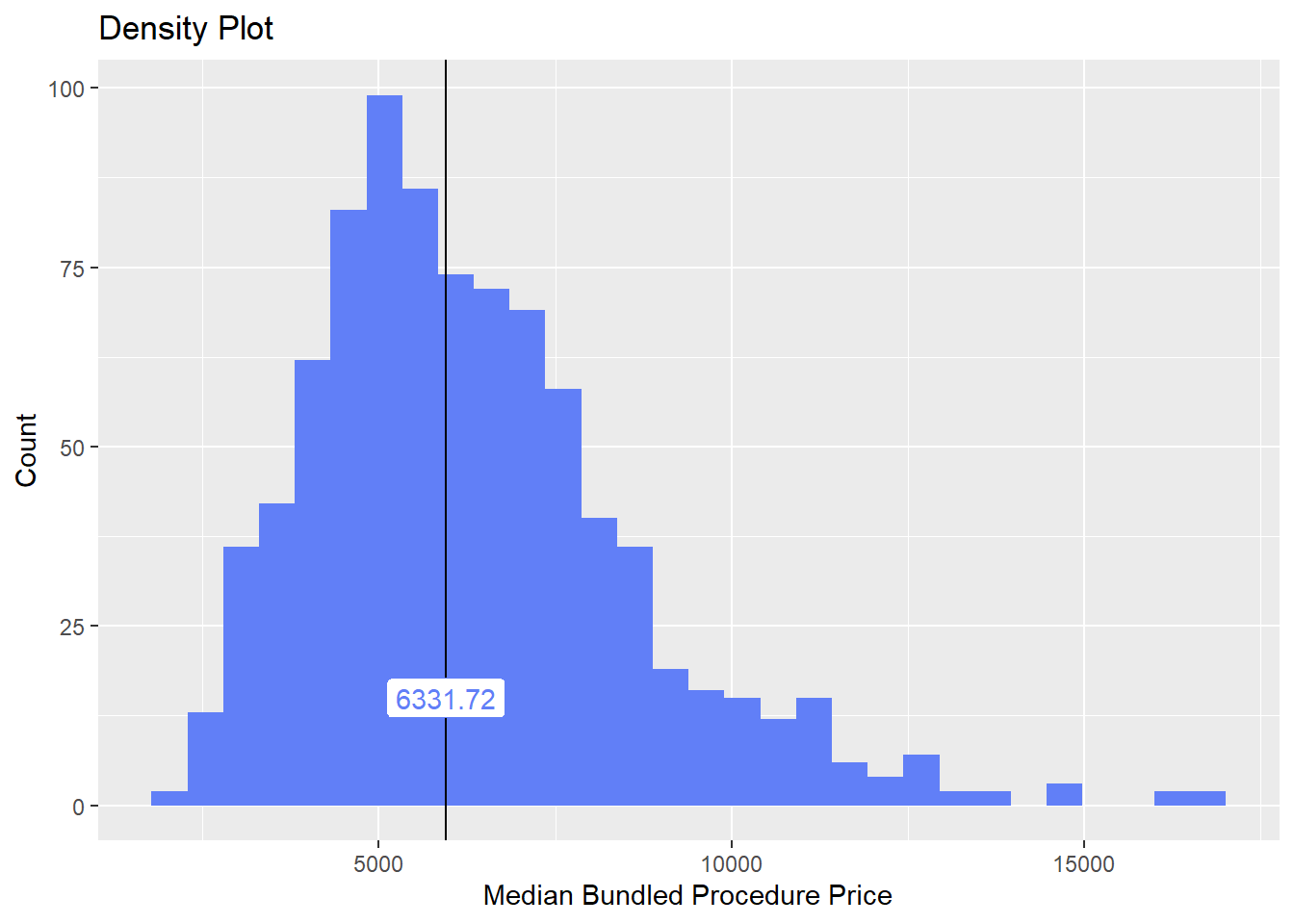
hernia_n_lap %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 166 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_hernia 0.276
## 2 surg_bun_t_repair 0.273
## 3 surg_bun_t_umbil 0.259
## 4 surg_bun_t_mesh 0.247
## 5 duration_max 0.121
## 6 medi_bun_t_oral 0.120
## 7 surg_bun_t_arm 0.0969
## 8 surg_bun_t_elbow 0.0969
## 9 anes_bun_t_head 0.0969
## 10 anes_bun_t_neck 0.0969
## # ... with 156 more rowshernia_n_lap %>% get_tag_density_information("medi_bun_t_oral") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ medi_bun_t_oral"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 477## $dist_plots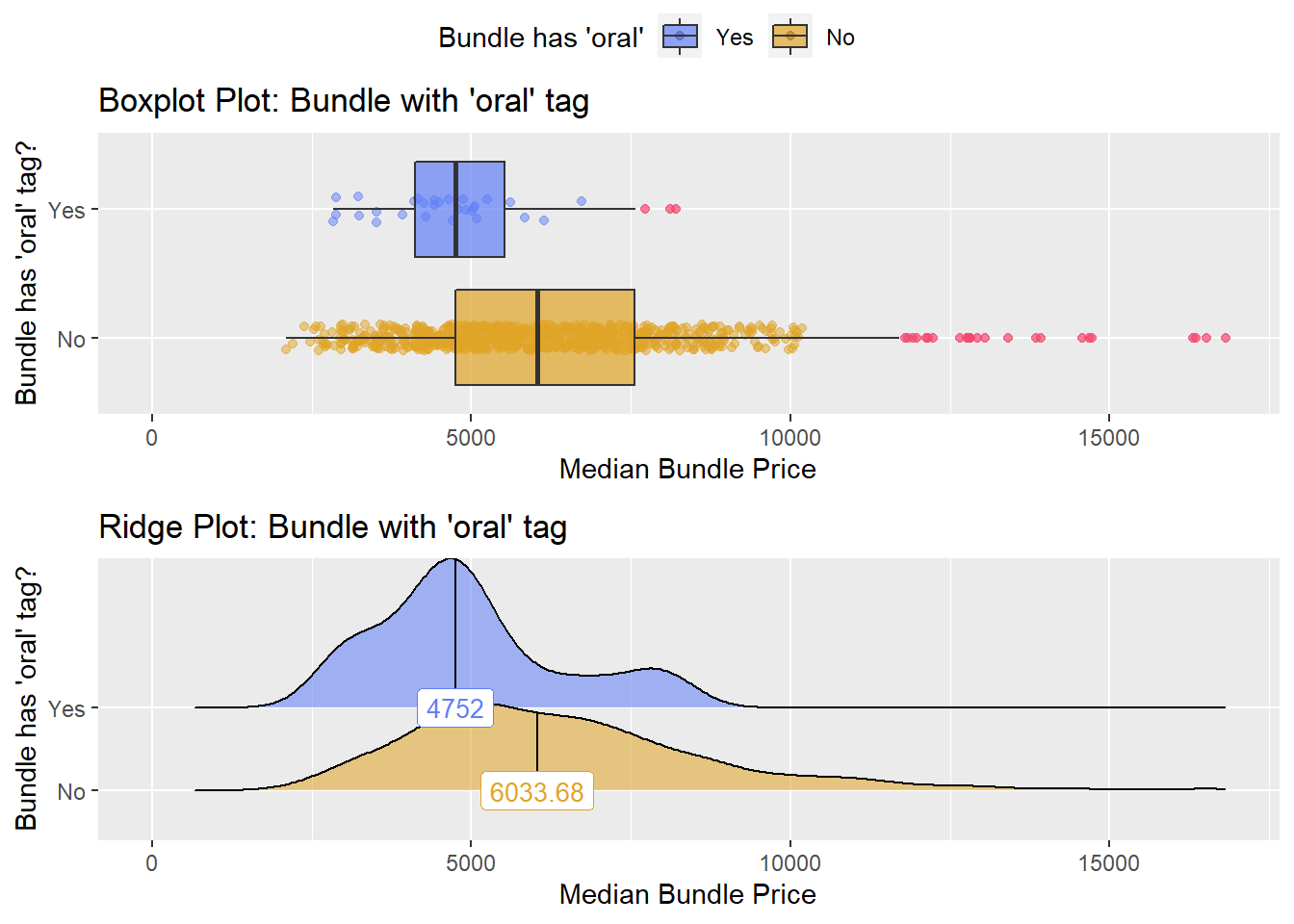
##
## $stat_tables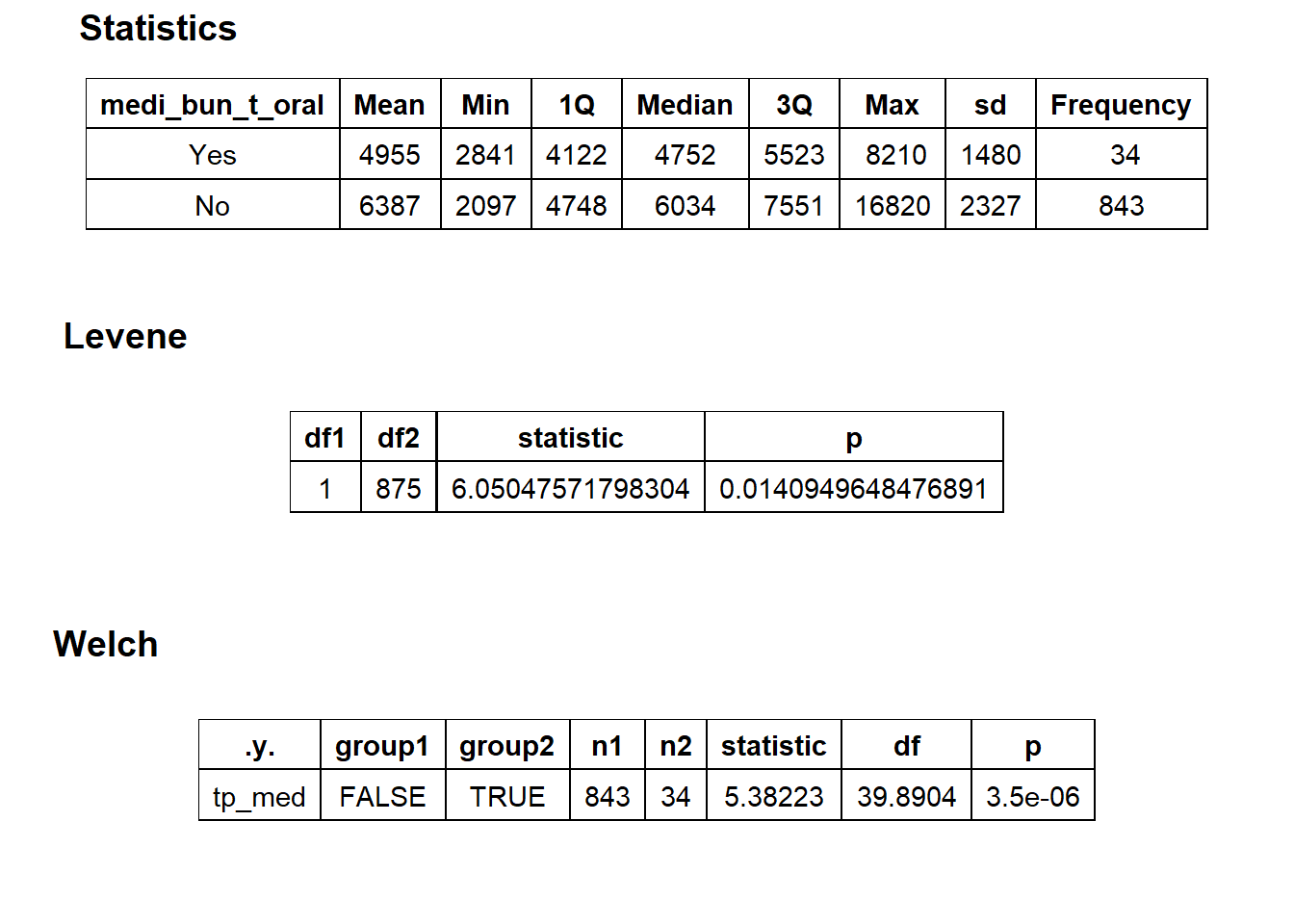
hernia_n_lap <- hernia_n_lap[medi_bun_t_oral ==F]hernia_n_lap %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.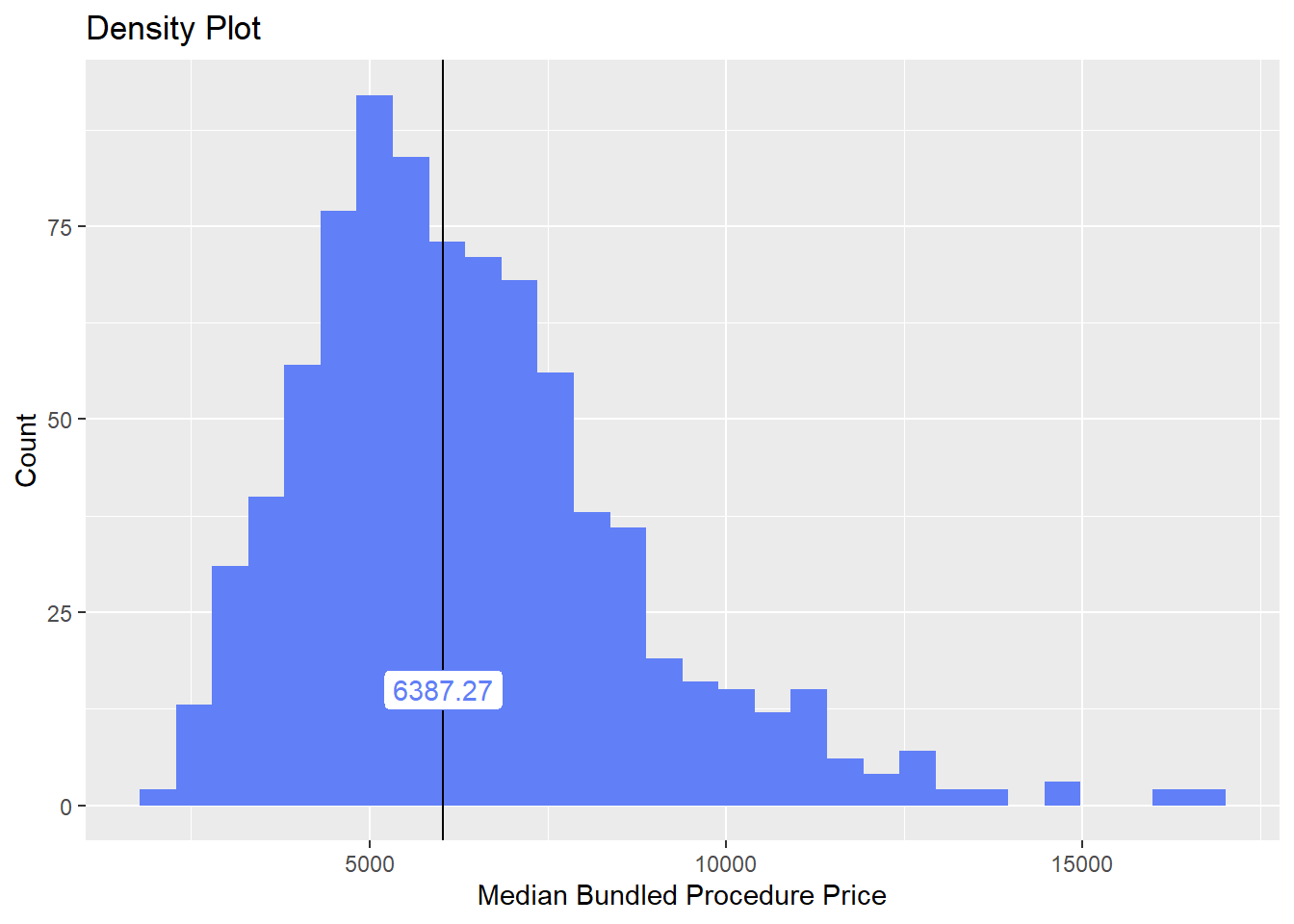
hernia_n_lap %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 161 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_hernia 0.275
## 2 surg_bun_t_repair 0.274
## 3 surg_bun_t_umbil 0.254
## 4 surg_bun_t_mesh 0.250
## 5 duration_max 0.121
## 6 surg_bun_t_arm 0.0972
## 7 surg_bun_t_elbow 0.0972
## 8 anes_bun_t_head 0.0972
## 9 anes_bun_t_neck 0.0972
## 10 medi_bun_t_tracing 0.0959
## # ... with 151 more rowshernia_n_lap_btbv4 <- hernia_n_lap %>% btbv4() %>% .[,
primary_doctor := pmap(.l=list(doctor_npi1=doctor_npi_str1,
doctor_npi2=doctor_npi_str2,
class_reqs="Surgery"#,
# specialization_reqs = ""
),
.f=calculate_primary_doctor) %>% as.character()
] %>%
#Filter out any procedures where our doctors fail both criteria.
.[!(primary_doctor %in% c("BOTH_DOC_FAIL_CRIT", "TWO_FIT_ALL_SPECS", "NONE_FIT_SPEC_REQ"))] %>%
.[,primary_doctor_npi := fifelse(primary_doctor==doctor_str1,
doctor_npi_str1,
doctor_npi_str2)] %>%
.[,`:=`(procedure_type=6, procedure_modifier="Hernia Repair")]## [1] "multiple meet class req"
## [1] "TREVOR DAY" "CAMERON TALMAGE BLACK"
## [1] "multiple meet class req"
## [1] "JOHN LARSEN CLAYTON" "ALICE FAITH YUO CHUNG"
## [1] "multiple meet class req"
## [1] "RODRICK D MCKINLAY" "STEVEN CHARLES SIMPER"
## [1] "multiple meet class req"
## [1] "TIMOTHY C HOLLINGSED" "ROBERT L RODRIGUES"
## [1] "multiple meet class req"
## [1] "MILDA SHAPIRO" "WILLIAM NOEL PEUGH"
## [1] "multiple meet class req"
## [1] "DANIELLE M. ADAMS" "VANESSA MARIE HART"
## [1] "multiple meet class req"
## [1] "ROBERT PATTERSON" "MILDA SHAPIRO"hernia_full <- hernia_lap_btbv4 %>% rbind(hernia_n_lap_btbv4)hernia_full %>% count(most_important_fac) %>% arrange(desc(n)) %>% View()hernia_bq <- hernia_full %>% .[,.(tp_med_med = median(tp_med_med),
tp_med_surg = median(tp_med_surg),
tp_med_medi = median(tp_med_medi),
tp_med_path = median(tp_med_path),
tp_med_radi = median(tp_med_radi),
tp_med_anes = median(tp_med_anes),
tp_med_faci = median(tp_med_faci),
tp_cnt_cnt = sum(tp_cnt_cnt)
),
by=c("procedure_type", "procedure_modifier","primary_doctor", "primary_doctor_npi", "most_important_fac", "most_important_fac_npi")] %>%
.[,.(
primary_doctor,
primary_doctor_npi,
most_important_fac ,
most_important_fac_npi,
procedure_type,
procedure_modifier,
tp_med_med,
tp_med_surg,
tp_med_medi,
tp_med_path,
tp_med_radi,
tp_med_anes,
tp_med_faci,
tp_cnt_cnt,
ingest_date = Sys.Date()
)]hernia_bq$tp_med_med %>% summary()## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 2612 6034 7784 8399 10392 19949# hernia_bq %>% fwrite("2020-12-17-hernia_repiar_fixed.csv")hernia_bq <- hernia_bq[,.(
primary_doctor,
primary_doctor_npi,
most_important_fac ,
most_important_fac_npi,
procedure_type,
procedure_modifier,
tp_med_med,
tp_med_surg,
tp_med_medi,
tp_med_path,
tp_med_radi,
tp_med_anes,
tp_med_faci,
ingest_date = Sys.Date()
)]bq_table_upload(x=procedure_table, values= hernia_bq, create_disposition='CREATE_IF_NEEDED', write_disposition='WRITE_APPEND')