Hip Replacement
Introduction
In this section we will:
* identify which Hip Replacement procedures were excluded and why
* identify which different Hip Replacement subgroups were created and why
If you have questions or concerns about this data please contact Alexander Nielson (alexnielson@utah.gov)
Load Libraries
Load Libraries
library(data.table)
library(tidyverse)## Warning: replacing previous import 'vctrs::data_frame' by 'tibble::data_frame'
## when loading 'dplyr'library(stringi)
library(ggridges)
library(broom)
library(disk.frame)
library(RecordLinkage)
library(googlesheets4)
library(bigrquery)
library(DBI)
devtools::install_github("utah-osa/hcctools2", upgrade="never" )
library(hcctools2)Establish color palettes
cust_color_pal1 <- c(
"Anesthesia" = "#f3476f",
"Facility" = "#e86a33",
"Medicine Proc" = "#e0a426",
"Pathology" = "#77bf45",
"Radiology" = "#617ff7",
"Surgery" = "#a974ee"
)
cust_color_pal2 <- c(
"TRUE" = "#617ff7",
"FALSE" = "#e0a426"
)
cust_color_pal3 <- c(
"above avg" = "#f3476f",
"avg" = "#77bf45",
"below avg" = "#a974ee"
)
fac_ref_regex <- "(UTAH)|(IHC)|(HOSP)|(HOSPITAL)|(CLINIC)|(ANESTH)|(SCOPY)|(SURG)|(LLC)|(ASSOC)|(MEDIC)|(CENTER)|(ASSOCIATES)|(U OF U)|(HEALTH)|(OLOGY)|(OSCOPY)|(FAMILY)|(VAMC)|(SLC)|(SALT LAKE)|(CITY)|(PROVO)|(OGDEN)|(ENDO )|( VALLEY)|( REGIONAL)|( CTR)|(GRANITE)|( INSTITUTE)|(INSTACARE)|(WASATCH)|(COUNTY)|(PEDIATRIC)|(CORP)|(CENTRAL)|(URGENT)|(CARE)|(UNIV)|(ODYSSEY)|(MOUNTAINSTAR)|( ORTHOPEDIC)|(INSTITUT)|(PARTNERSHIP)|(PHYSICIAN)|(CASTLEVIEW)|(CONSULTING)|(MAGEMENT)|(PRACTICE)|(EMERGENCY)|(SPECIALISTS)|(DIVISION)|(GUT WHISPERER)|(INTERMOUNTAIN)|(OBGYN)"Connect to GCP database
bigrquery::bq_auth(path = 'D:/gcp_keys/healthcare-compare-prod-95b3b7349c32.json')
# set my project ID and dataset name
project_id <- 'healthcare-compare-prod'
dataset_name <- 'healthcare_compare'
con <- dbConnect(
bigrquery::bigquery(),
project = project_id,
dataset = dataset_name,
billing = project_id
)Get NPI table
query <- paste0("SELECT npi, clean_name, osa_group, osa_class, osa_specialization
FROM `healthcare-compare-prod.healthcare_compare.npi_full`")
#bq_project_query(billing, query) # uncomment to determine billing price for above query.
npi_full <- dbGetQuery(con, query) %>%
data.table() get a subset of the NPI providers based upon taxonomy groups
gs4_auth(email="alexnielson@utah.gov")surgery <- read_sheet("https://docs.google.com/spreadsheets/d/1GY8lKwUJuPHtpUl4EOw9eriLUDG9KkNWrMbaSnA5hOU/edit#gid=0",
sheet="major_surgery") %>% as.data.table## Reading from "Doctor Types to Keep"## Range "'major_surgery'"surgery <- surgery[is.na(Remove) ] %>% .[["NUCC Classification"]] npi_prov_pair <- npi_full[osa_class %in% surgery] %>%
.[,.(npi=npi,
clean_name = clean_name
)
] Load Data
bun_proc <- disk.frame("full_apcd.df")hip <- bun_proc[surg_bun_t_hip==T & surg_bun_t_arthro==T]hip <- hip[,`:=`(
surg_sp_name_clean = surg_sp_npi %>% map_chr(get_npi_standard_name),
surg_bp_name_clean = surg_bp_npi %>% map_chr(get_npi_standard_name),
medi_sp_name_clean = medi_sp_npi %>% map_chr(get_npi_standard_name),
medi_bp_name_clean = medi_bp_npi %>% map_chr(get_npi_standard_name),
radi_sp_name_clean = radi_sp_npi %>% map_chr(get_npi_standard_name),
radi_bp_name_clean = radi_bp_npi %>% map_chr(get_npi_standard_name),
path_sp_name_clean = path_sp_npi %>% map_chr(get_npi_standard_name),
path_bp_name_clean = path_bp_npi %>% map_chr(get_npi_standard_name),
anes_sp_name_clean = anes_sp_npi %>% map_chr(get_npi_standard_name),
anes_bp_name_clean = anes_bp_npi %>% map_chr(get_npi_standard_name),
faci_sp_name_clean = faci_sp_npi %>% map_chr(get_npi_standard_name),
faci_bp_name_clean = faci_bp_npi %>% map_chr(get_npi_standard_name)
)]hip %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
hip %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 255 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_arthroplasty 0.309
## 2 surg_bun_t_total 0.309
## 3 duration_max 0.263
## 4 radi_bun_t_pelvi 0.246
## 5 radi_bun_t_exam 0.233
## 6 radi_bun_t_x-ray 0.233
## 7 anes_bun_t_anes 0.202
## 8 anes_bun_t_anesth 0.202
## 9 surg_bun_t_repair 0.201
## 10 medi_bun_t_inject 0.185
## # ... with 245 more rowship %>% get_tag_density_information("surg_bun_t_arthroplasty") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_arthroplasty"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 2350## $dist_plots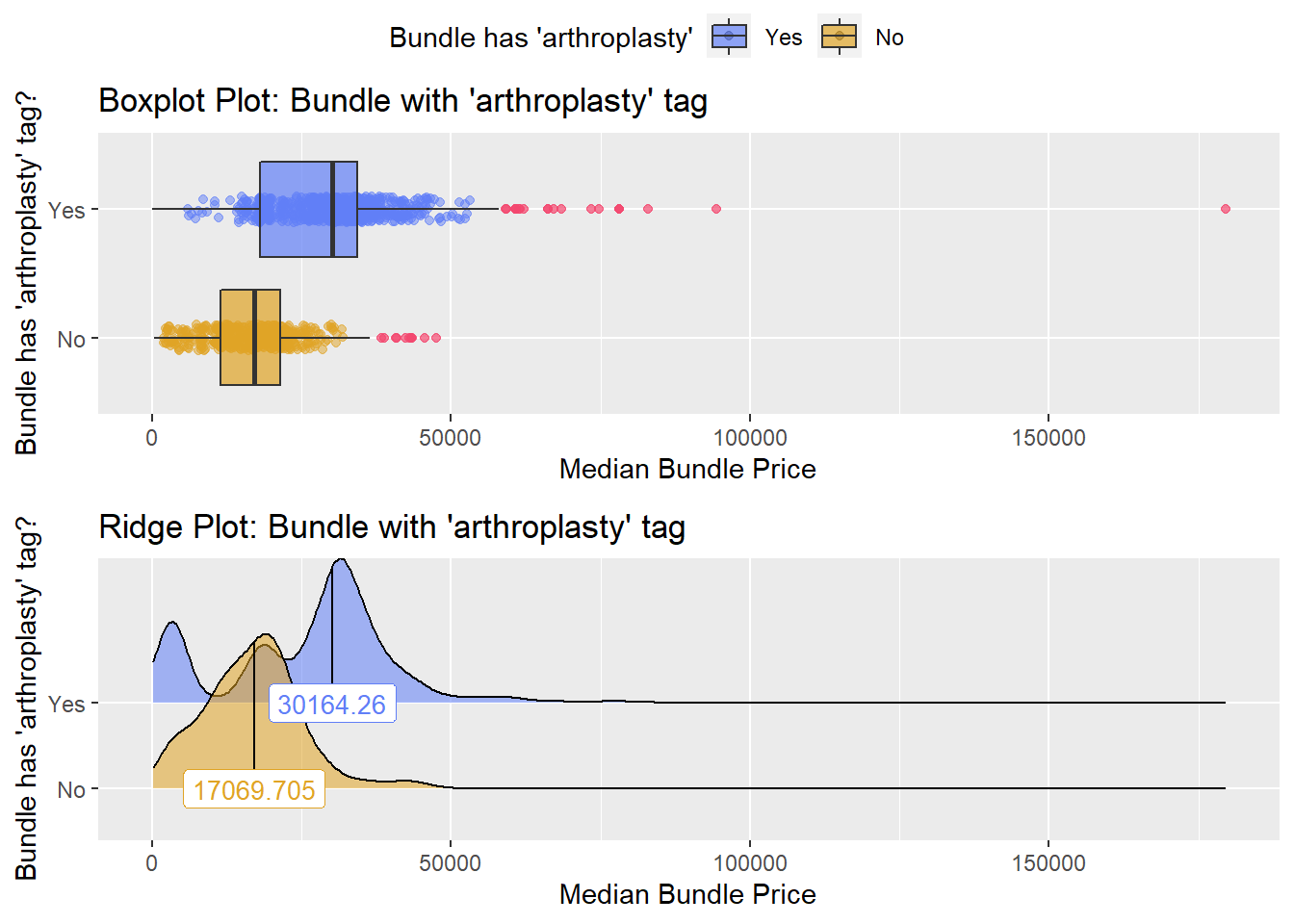
##
## $stat_tables
hip_total <- hip[surg_bun_t_arthroplasty==T & cnt>2 & faci_bun_sum_med > 500]
hip_scopy <- hip[surg_bun_t_arthroplasty==F]work on total hip arhtroplasties first
once again check price density and correlated tags
hip_total %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
hip_total %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 200 x 2
## name correlation
## <chr> <dbl>
## 1 radi_bun_t_without_dye 0.284
## 2 surg_bun_t_place 0.278
## 3 surg_bun_t_revise 0.278
## 4 radi_bun_t_ct 0.272
## 5 anes_bun_t_surg 0.250
## 6 anes_bun_t_joint 0.242
## 7 surg_bun_t_resect 0.236
## 8 surg_bun_t_joint 0.219
## 9 radi_bun_t_arm 0.207
## 10 radi_bun_t_leg 0.207
## # ... with 190 more rowship_total %>% get_tag_density_information("radi_bun_t_without_dye") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ radi_bun_t_without_dye"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 12400## $dist_plots
##
## $stat_tables
hip_total <- hip_total[radi_bun_t_without_dye==F]once again check price density and correlated tags
hip_total %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
hip_total %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 157 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_embolize 0.176
## 2 surg_bun_t_resect 0.176
## 3 radi_bun_t_arm 0.176
## 4 radi_bun_t_artery 0.176
## 5 radi_bun_t_leg 0.176
## 6 radi_bun_t_vessel 0.176
## 7 anes_bun_t_mod_sed 0.176
## 8 surg_bun_t_pelvis 0.169
## 9 surg_bun_t_exc 0.151
## 10 surg_bun_t_tum 0.140
## # ... with 147 more rows# hip_total %>% get_tag_density_information("surg_bun_t_resect") %>% print()
hip_total[surg_bun_t_resect ==T] %>% nrow()## [1] 1there is only one with this so we will omit
hip_total <- hip_total[surg_bun_t_resect==F]once again check price density and correlated tags
hip_total %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
hip_total %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 150 x 2
## name correlation
## <chr> <dbl>
## 1 surg_bun_t_pelvis 0.172
## 2 anes_bun_t_anes 0.166
## 3 anes_bun_t_anesth 0.166
## 4 surg_bun_t_exc 0.154
## 5 anes_bun_t_repair 0.143
## 6 path_bun_t_hormone 0.129
## 7 path_bun_t_thyroid 0.129
## 8 surg_bun_t_place 0.118
## 9 surg_bun_t_revise 0.118
## 10 surg_bun_t_dev 0.0988
## # ... with 140 more rowship_total %>% get_tag_density_information("surg_bun_t_pelvis") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ surg_bun_t_pelvis"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 785## $dist_plots
##
## $stat_tables
There are only two, so they do not need to be their own option in the tool.
hip_total <- hip_total[surg_bun_t_pelvis==F]once again check price density and correlated tags
hip_total %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
hip_total %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 149 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_anes 0.167
## 2 anes_bun_t_anesth 0.167
## 3 anes_bun_t_repair 0.146
## 4 path_bun_t_hormone 0.131
## 5 path_bun_t_thyroid 0.131
## 6 surg_bun_t_place 0.121
## 7 surg_bun_t_revise 0.121
## 8 surg_bun_t_dev 0.101
## 9 surg_bun_t_device 0.101
## 10 surg_bun_t_drug 0.101
## # ... with 139 more rowship_total %>% get_tag_density_information("anes_bun_t_anes") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ anes_bun_t_anes"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 2990## $dist_plots
##
## $stat_tables
I am really only interested in the proceduers which included anesthesia costs for the total hip replacement.
hip_total <- hip_total[anes_bun_t_anes == T]once again check price density and correlated tags
hip_total %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
hip_total %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 141 x 2
## name correlation
## <chr> <dbl>
## 1 anes_bun_t_repair 0.15
## 2 path_bun_t_hormone 0.137
## 3 path_bun_t_thyroid 0.137
## 4 surg_bun_t_place 0.125
## 5 surg_bun_t_revise 0.125
## 6 duration_mean 0.118
## 7 surg_bun_t_joint 0.113
## 8 radi_bun_t_exam 0.105
## 9 radi_bun_t_x-ray 0.105
## 10 surg_bun_t_dev 0.104
## # ... with 131 more rowship_total %>% get_tag_density_information("anes_bun_t_repair") %>% print()## Scale for 'y' is already present. Adding another scale for 'y', which will
## replace the existing scale.## Scale for 'x' is already present. Adding another scale for 'x', which will
## replace the existing scale.## [1] "tp_med ~ anes_bun_t_repair"## Warning in leveneTest.default(y = y, group = group, ...): group coerced to
## factor.## Picking joint bandwidth of 3320## $dist_plots
##
## $stat_tables
hip_total <- hip_total[anes_bun_t_repair==F]once again check price density and correlated tags
hip_total %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
hip_total %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 136 x 2
## name correlation
## <chr> <dbl>
## 1 path_bun_t_hormone 0.140
## 2 path_bun_t_thyroid 0.140
## 3 duration_mean 0.113
## 4 radi_bun_t_exam 0.106
## 5 radi_bun_t_x-ray 0.106
## 6 radi_bun_t_pelvi 0.0961
## 7 radi_bun_t_chest 0.082
## 8 faci_bun_t_consultation 0.0809
## 9 faci_bun_t_inpatient 0.0809
## 10 medi_bun_t_inject 0.0766
## # ... with 126 more rows# hip_total %>% get_tag_density_information("path_bun_t_thyroid") %>% print()
hip_total[path_bun_t_thyroid==T] %>% nrow()## [1] 1only one so we will omit it.
hip_total <- hip_total[path_bun_t_thyroid==F]once again check price density and correlated tags
hip_total %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
hip_total %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 134 x 2
## name correlation
## <chr> <dbl>
## 1 duration_mean 0.104
## 2 radi_bun_t_exam 0.0987
## 3 radi_bun_t_x-ray 0.0987
## 4 radi_bun_t_pelvi 0.0903
## 5 radi_bun_t_chest 0.0833
## 6 faci_bun_t_consultation 0.0819
## 7 faci_bun_t_inpatient 0.0819
## 8 medi_bun_t_inject 0.0766
## 9 path_bun_t_blood 0.0765
## 10 radi_bun_t_angiography 0.0737
## # ... with 124 more rows# hip_total %>% get_tag_density_information("faci_bun_t_consultation") %>% print()
hip_total[faci_bun_t_consultation==T] %>% nrow()## [1] 1hip_total <- hip_total[faci_bun_t_consultation==F]once again check price density and correlated tags
hip_total %>% plot_med_density() %>% print()## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
hip_total %>% get_tag_cor() %>% print()## Warning in stats::cor(cor_data): the standard deviation is zero## # A tibble: 132 x 2
## name correlation
## <chr> <dbl>
## 1 duration_mean 0.103
## 2 radi_bun_t_exam 0.094
## 3 radi_bun_t_x-ray 0.094
## 4 radi_bun_t_pelvi 0.0868
## 5 radi_bun_t_chest 0.0838
## 6 path_bun_t_blood 0.0765
## 7 medi_bun_t_inject 0.0765
## 8 radi_bun_t_angiography 0.0741
## 9 radi_bun_t_ct 0.0741
## 10 path_bun_t_glucose 0.0699
## # ... with 122 more rowship_total_btbv4 <- hip_total %>% btbv4()hip_total_bq <- hip_total_btbv4[,
primary_doctor := pmap(.l=list(doctor_npi1=doctor_npi_str1,
doctor_npi2=doctor_npi_str2,
class_reqs="Orthopaedic Surgery"
),
.f=calculate_primary_doctor) %>% as.character()
] %>%
#Filter out any procedures where our doctors fail both criteria.
.[!(primary_doctor %in% c("BOTH_DOC_FAIL_CRIT", "TWO_FIT_ALL_SPECS"))] %>%
.[,primary_doctor_npi := fifelse(primary_doctor==doctor_str1,
doctor_npi_str1,
doctor_npi_str2)] %>%
.[,`:=`(procedure_type=3, procedure_modifier="Total Hip Replacement")]## [1] "multiple meet class req"
## [1] "CHRISTOPHER EARL PELT" "MICHAEL CHAD MAHAN"
## [1] "multiple meet class req"
## [1] "JEREMY MARK GILILLAND" "MICHAEL CHAD MAHAN"
## [1] "multiple meet class req"
## [1] "JEREMY MARK GILILLAND" "KEVIN CAMPBELL"
## [1] "multiple meet class req"
## [1] "CHRISTOPHER L. PETERS" "KEVIN CAMPBELL"
## [1] "multiple meet class req"
## [1] "CHRISTOPHER L. PETERS" "STEVEN DONOHOE"
## [1] "multiple meet class req"
## [1] "CHRISTOPHER EARL PELT" "STEVEN DONOHOE"
## [1] "multiple meet class req"
## [1] "CHRISTOPHER L. PETERS" "MICHAEL CHAD MAHAN"
## [1] "multiple meet class req"
## [1] "MICHAEL H BOURNE" "E MARC MARIANI"
## [1] "multiple meet class req"
## [1] "DARIN KIMBALL ALLRED" "WARREN LEWIS BUTTERFIELD"hip_total_bq <- hip_total_bq[,.(
primary_doctor,
primary_doctor_npi,
most_important_fac ,
most_important_fac_npi,
procedure_type,
procedure_modifier,
tp_med_med,
tp_med_surg,
tp_med_medi,
tp_med_path,
tp_med_radi,
tp_med_anes,
tp_med_faci
)]bq_table_upload(x=procedure_table, values= hip_total_bq, create_disposition='CREATE_IF_NEEDED', write_disposition='WRITE_APPEND')